Next-generation sequencing of human mitochondrial reference genomes uncovers high heteroplasmy frequency
- PMID: 23133345
- PMCID: PMC3486893
- DOI: 10.1371/journal.pcbi.1002737
Next-generation sequencing of human mitochondrial reference genomes uncovers high heteroplasmy frequency
Abstract
We describe methods for rapid sequencing of the entire human mitochondrial genome (mtgenome), which involve long-range PCR for specific amplification of the mtgenome, pyrosequencing, quantitative mapping of sequence reads to identify sequence variants and heteroplasmy, as well as de novo sequence assembly. These methods have been used to study 40 publicly available HapMap samples of European (CEU) and African (YRI) ancestry to demonstrate a sequencing error rate <5.63×10(-4), nucleotide diversity of 1.6×10(-3) for CEU and 3.7×10(-3) for YRI, patterns of sequence variation consistent with earlier studies, but a higher rate of heteroplasmy varying between 10% and 50%. These results demonstrate that next-generation sequencing technologies allow interrogation of the mitochondrial genome in greater depth than previously possible which may be of value in biology and medicine.
Conflict of interest statement
SM: I have read the journal's policy and have the following conflict: Certain commercial equipment or materials are identified in this report to specify adequately the experimental procedures. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials or equipment identified are necessarily the best available for the purpose.
Figures
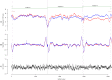
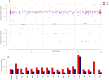

References
-
- Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, et al. (1981) Sequence and organization of the human mitochondrial genome. Nature 290: 457–465. - PubMed
-
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, et al. (1999) Reanalysis and revision of the cambridge reference sequence for human mitochondrial DNA. Nat Genet 23: 147. - PubMed
-
- Hutchinson CA, Newbold JE, Potter SS, Edgell MH (1974) Maternal inheritance of mammalian mitochondrial DNA. Nature 251: 536–538. - PubMed
-
- Olivo PD, Van de Walle, Michale J, Laipist PJ, Hauswirth WW (1983) Nucleotide sequence evidence for rapid genotypic shifts in the bovine mitochondrial DNA D-loop. Nature 306: 400. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

