Comparative analysis of RNA families reveals distinct repertoires for each domain of life
- PMID: 23133357
- PMCID: PMC3486863
- DOI: 10.1371/journal.pcbi.1002752
Comparative analysis of RNA families reveals distinct repertoires for each domain of life
Abstract
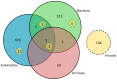
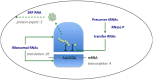
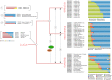
The RNA world hypothesis, that RNA genomes and catalysts preceded DNA genomes and genetically-encoded protein catalysts, has been central to models for the early evolution of life on Earth. A key part of such models is continuity between the earliest stages in the evolution of life and the RNA repertoires of extant lineages. Some assessments seem consistent with a diverse RNA world, yet direct continuity between modern RNAs and an RNA world has not been demonstrated for the majority of RNA families, and, anecdotally, many RNA functions appear restricted in their distribution. Despite much discussion of the possible antiquity of RNA families, no systematic analyses of RNA family distribution have been performed. To chart the broad evolutionary history of known RNA families, we performed comparative genomic analysis of over 3 million RNA annotations spanning 1446 families from the Rfam 10 database. We report that 99% of known RNA families are restricted to a single domain of life, revealing discrete repertoires for each domain. For the 1% of RNA families/clans present in more than one domain, over half show evidence of horizontal gene transfer (HGT), and the rest show a vertical trace, indicating the presence of a complex protein synthesis machinery in the Last Universal Common Ancestor (LUCA) and consistent with the evolutionary history of the most ancient protein-coding genes. However, with limited interdomain transfer and few RNA families exhibiting demonstrable antiquity as predicted under RNA world continuity, our results indicate that the majority of modern cellular RNA repertoires have primarily evolved in a domain-specific manner.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
The evolutionary history of protein fold families and proteomes confirms that the archaeal ancestor is more ancient than the ancestors of other superkingdoms.BMC Evol Biol. 2012 Jan 27;12:13. doi: 10.1186/1471-2148-12-13. BMC Evol Biol. 2012. PMID: 22284070 Free PMC article.
-
Comparative genomics of eukaryotic small nucleolar RNAs reveals deep evolutionary ancestry amidst ongoing intragenomic mobility.BMC Evol Biol. 2012 Sep 15;12:183. doi: 10.1186/1471-2148-12-183. BMC Evol Biol. 2012. PMID: 22978381 Free PMC article.
-
The last universal common ancestor between ancient Earth chemistry and the onset of genetics.PLoS Genet. 2018 Aug 16;14(8):e1007518. doi: 10.1371/journal.pgen.1007518. eCollection 2018 Aug. PLoS Genet. 2018. PMID: 30114187 Free PMC article. Review.
-
Ribonucleotide reduction - horizontal transfer of a required function spans all three domains.BMC Evol Biol. 2010 Dec 10;10:383. doi: 10.1186/1471-2148-10-383. BMC Evol Biol. 2010. PMID: 21143941 Free PMC article.
-
Archaea-First and the Co-Evolutionary Diversification of Domains of Life.Bioessays. 2018 Aug;40(8):e1800036. doi: 10.1002/bies.201800036. Epub 2018 Jun 26. Bioessays. 2018. PMID: 29944192 Review.
Cited by
-
Rfam 11.0: 10 years of RNA families.Nucleic Acids Res. 2013 Jan;41(Database issue):D226-32. doi: 10.1093/nar/gks1005. Epub 2012 Nov 3. Nucleic Acids Res. 2013. PMID: 23125362 Free PMC article.
-
The very early evolution of protein translocation across membranes.PLoS Comput Biol. 2021 Mar 8;17(3):e1008623. doi: 10.1371/journal.pcbi.1008623. eCollection 2021 Mar. PLoS Comput Biol. 2021. PMID: 33684113 Free PMC article.
-
The natural history of transfer RNA and its interactions with the ribosome.Front Genet. 2014 May 9;5:127. doi: 10.3389/fgene.2014.00127. eCollection 2014. Front Genet. 2014. PMID: 24847358 Free PMC article. No abstract available.
-
In silico ribozyme evolution in a metabolically coupled RNA population.Biol Direct. 2015 May 27;10:30. doi: 10.1186/s13062-015-0049-6. Biol Direct. 2015. PMID: 26014147 Free PMC article.
-
Diversity and prevalence of ANTAR RNAs across actinobacteria.BMC Microbiol. 2021 May 29;21(1):159. doi: 10.1186/s12866-021-02234-x. BMC Microbiol. 2021. PMID: 34051745 Free PMC article.
References
-
- Fraenkel-Conrat H (1956) The role of the nucleic acid in the reconstitution of active Tobacco Mosaic Virus. Journal of the American Chemical Society 78: 882–883.
-
- Gierer A, Schramm G (1956) Infectivity of ribonucleic acid from Tobacco Mosaic Virus. Nature 177: 702–703. - PubMed
-
- Diener TO (1971) Potato spindle tuber “virus”. IV. A replicating, low molecular weight RNA. Virology 45: 411–428. - PubMed
-
- Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, et al. (1982) Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31: 147–157. - PubMed
-
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S (1983) The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35: 849–857. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

