Transcription factor Amr1 induces melanin biosynthesis and suppresses virulence in Alternaria brassicicola
- PMID: 23133370
- PMCID: PMC3486909
- DOI: 10.1371/journal.ppat.1002974
Transcription factor Amr1 induces melanin biosynthesis and suppresses virulence in Alternaria brassicicola
Abstract
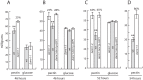
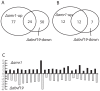
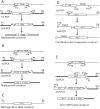
Alternaria brassicicola is a successful saprophyte and necrotrophic plant pathogen. Several A. brassicicola genes have been characterized as affecting pathogenesis of Brassica species. To study regulatory mechanisms of pathogenesis, we mined 421 genes in silico encoding putative transcription factors in a machine-annotated, draft genome sequence of A. brassicicola. In this study, targeted gene disruption mutants for 117 of the transcription factor genes were produced and screened. Three of these genes were associated with pathogenesis. Disruption mutants of one gene (AbPacC) were nonpathogenic and another gene (AbVf8) caused lesions less than half the diameter of wild-type lesions. Unexpectedly, mutants of the third gene, Amr1, caused lesions with a two-fold larger diameter than the wild type and complementation mutants. Amr1 is a homolog of Cmr1, a transcription factor that regulates melanin biosynthesis in several fungi. We created gene deletion mutants of Δamr1 and characterized their phenotypes. The Δamr1 mutants used pectin as a carbon source more efficiently than the wild type, were melanin-deficient, and more sensitive to UV light and glucanase digestion. The AMR1 protein was localized in the nuclei of hyphae and in highly melanized conidia during the late stage of plant pathogenesis. RNA-seq analysis revealed that three genes in the melanin biosynthesis pathway, along with the deleted Amr1 gene, were expressed at low levels in the mutants. In contrast, many hydrolytic enzyme-coding genes were expressed at higher levels in the mutants than in the wild type during pathogenesis. The results of this study suggested that a gene important for survival in nature negatively affected virulence, probably by a less efficient use of plant cell-wall materials. We speculate that the functions of the Amr1 gene are important to the success of A. brassicicola as a competitive saprophyte and plant parasite.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Churchill A, LU S, Turgeon B, Yoder O, Macko V (1995) Victorin-deficient REMI mutants of Cochliobolus victoriae demonstrate a requirement for victorin in pathogenesis. Fungal Genet Newslett 42A: 41.
-
- Del Sorbo G, Scala F, Parrella G, Lorito M, Comparini C, et al. (2000) Functional expression of the gene cu, encoding the phytotoxic hydrophobin cerato-ulmin, enables Ophiostoma quercus, a nonpathogen on elm, to cause symptoms of Dutch elm disease. Mol Plant Microbe Interact 13: 43–53. - PubMed
-
- Yun S-H, Turgeon B, Yoder O (1998) REM-induced mutants of Mycosphaerella zeae-maydis lacking the polyketide PM-toxin are deficient in pathogenesis to corn. Physiol Mol Plant Pathol 52: 53–66.
-
- Ito K, Tanaka T, Hatta R, Yamamoto M, Akimitsu K, et al. (2004) Dissection of the host range of the fungal plant pathogen Alternaria alternata by modification of secondary metabolism. Mol Microbiol 52: 399–411. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

