Histone H1 plays a role in heterochromatin formation and VSG expression site silencing in Trypanosoma brucei
- PMID: 23133390
- PMCID: PMC3486875
- DOI: 10.1371/journal.ppat.1003010
Histone H1 plays a role in heterochromatin formation and VSG expression site silencing in Trypanosoma brucei
Abstract
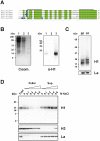
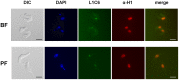
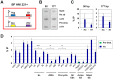
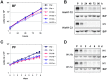
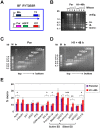
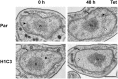
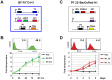
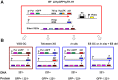
The African sleeping sickness parasite Trypanosoma brucei evades the host immune system through antigenic variation of its variant surface glycoprotein (VSG) coat. Although the T. brucei genome contains ∼1500 VSGs, only one VSG is expressed at a time from one of about 15 subtelomeric VSG expression sites (ESs). For antigenic variation to work, not only must the vast VSG repertoire be kept silent in a genome that is mainly constitutively transcribed, but the frequency of VSG switching must be strictly controlled. Recently it has become clear that chromatin plays a key role in silencing inactive ESs, thereby ensuring monoallelic expression of VSG. We investigated the role of the linker histone H1 in chromatin organization and ES regulation in T. brucei. T. brucei histone H1 proteins have a different domain structure to H1 proteins in higher eukaryotes. However, we show that they play a key role in the maintenance of higher order chromatin structure in bloodstream form T. brucei as visualised by electron microscopy. In addition, depletion of histone H1 results in chromatin becoming generally more accessible to endonucleases in bloodstream but not in insect form T. brucei. The effect on chromatin following H1 knock-down in bloodstream form T. brucei is particularly evident at transcriptionally silent ES promoters, leading to 6-8 fold derepression of these promoters. T. brucei histone H1 therefore appears to be important for the maintenance of repressed chromatin in bloodstream form T. brucei. In particular H1 plays a role in downregulating silent ESs, arguing that H1-mediated chromatin functions in antigenic variation in T. brucei.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Active VSG expression sites in Trypanosoma brucei are depleted of nucleosomes.Eukaryot Cell. 2010 Jan;9(1):136-47. doi: 10.1128/EC.00281-09. Epub 2009 Nov 13. Eukaryot Cell. 2010. PMID: 19915073 Free PMC article.
-
The FACT subunit TbSpt16 is involved in cell cycle specific control of VSG expression sites in Trypanosoma brucei.Mol Microbiol. 2010 Oct;78(2):459-74. doi: 10.1111/j.1365-2958.2010.07350.x. Epub 2010 Sep 2. Mol Microbiol. 2010. PMID: 20879999 Free PMC article.
-
Histone deacetylases play distinct roles in telomeric VSG expression site silencing in African trypanosomes.Mol Microbiol. 2010 Sep;77(5):1237-45. doi: 10.1111/j.1365-2958.2010.07284.x. Mol Microbiol. 2010. PMID: 20624217 Free PMC article.
-
Regulation of vsg expression site transcription and switching in Trypanosoma brucei.Mol Biochem Parasitol. 1998 Mar 1;91(1):77-91. doi: 10.1016/s0166-6851(97)00186-2. Mol Biochem Parasitol. 1998. PMID: 9574927 Review.
-
African trypanosomes: the genome and adaptations for immune evasion.Essays Biochem. 2011;51:47-62. doi: 10.1042/bse0510047. Essays Biochem. 2011. PMID: 22023441 Review.
Cited by
-
May the Odds Be Ever in Your Favor: Non-deterministic Mechanisms Diversifying Cell Surface Molecule Expression.Front Cell Dev Biol. 2022 Jan 11;9:720798. doi: 10.3389/fcell.2021.720798. eCollection 2021. Front Cell Dev Biol. 2022. PMID: 35087825 Free PMC article. Review.
-
Characterization of two different Asf1 histone chaperones with distinct cellular localizations and functions in Trypanosoma brucei.Nucleic Acids Res. 2014 Mar;42(5):2906-18. doi: 10.1093/nar/gkt1267. Epub 2013 Dec 9. Nucleic Acids Res. 2014. PMID: 24322299 Free PMC article.
-
Antigenic variation in African trypanosomes: the importance of chromosomal and nuclear context in VSG expression control.Cell Microbiol. 2013 Dec;15(12):1984-93. doi: 10.1111/cmi.12215. Epub 2013 Oct 10. Cell Microbiol. 2013. PMID: 24047558 Free PMC article. Review.
-
Trypanosoma brucei ATR Links DNA Damage Signaling during Antigenic Variation with Regulation of RNA Polymerase I-Transcribed Surface Antigens.Cell Rep. 2020 Jan 21;30(3):836-851.e5. doi: 10.1016/j.celrep.2019.12.049. Cell Rep. 2020. PMID: 31968257 Free PMC article.
-
TDP1 is an HMG chromatin protein facilitating RNA polymerase I transcription in African trypanosomes.Nucleic Acids Res. 2013 Mar 1;41(5):2981-92. doi: 10.1093/nar/gks1469. Epub 2013 Jan 29. Nucleic Acids Res. 2013. PMID: 23361461 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

