Population genomic scan for candidate signatures of balancing selection to guide antigen characterization in malaria parasites
- PMID: 23133397
- PMCID: PMC3486833
- DOI: 10.1371/journal.pgen.1002992
Population genomic scan for candidate signatures of balancing selection to guide antigen characterization in malaria parasites
Abstract
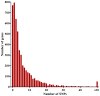
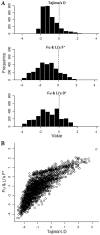
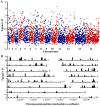
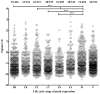
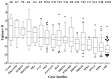
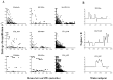
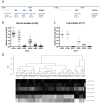
Acquired immunity in vertebrates maintains polymorphisms in endemic pathogens, leading to identifiable signatures of balancing selection. To comprehensively survey for genes under such selection in the human malaria parasite Plasmodium falciparum, we generated paired-end short-read sequences of parasites in clinical isolates from an endemic Gambian population, which were mapped to the 3D7 strain reference genome to yield high-quality genome-wide coding sequence data for 65 isolates. A minority of genes did not map reliably, including the hypervariable var, rifin, and stevor families, but 5,056 genes (90.9% of all in the genome) had >70% sequence coverage with minimum read depth of 5 for at least 50 isolates, of which 2,853 genes contained 3 or more single nucleotide polymorphisms (SNPs) for analysis of polymorphic site frequency spectra. Against an overall background of negatively skewed frequencies, as expected from historical population expansion combined with purifying selection, the outlying minority of genes with signatures indicating exceptionally intermediate frequencies were identified. Comparing genes with different stage-specificity, such signatures were most common in those with peak expression at the merozoite stage that invades erythrocytes. Members of clag, PfMC-2TM, surfin, and msp3-like gene families were highly represented, the strongest signature being in the msp3-like gene PF10_0355. Analysis of msp3-like transcripts in 45 clinical and 11 laboratory adapted isolates grown to merozoite-containing schizont stages revealed surprisingly low expression of PF10_0355. In diverse clonal parasite lines the protein product was expressed in a minority of mature schizonts (<1% in most lines and ∼10% in clone HB3), and eight sub-clones of HB3 cultured separately had an intermediate spectrum of positive frequencies (0.9 to 7.5%), indicating phase variable expression of this polymorphic antigen. This and other identified targets of balancing selection are now prioritized for functional study.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Grossman SR, Shylakhter I, Karlsson EK, Byrne EH, Morales S, et al. (2010) A composite of multiple signals distinguishes causal variants in regions of positive selection. Science 327: 883–886. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

