A dominantly acting murine allele of Mcm4 causes chromosomal abnormalities and promotes tumorigenesis
- PMID: 23133403
- PMCID: PMC3486839
- DOI: 10.1371/journal.pgen.1003034
A dominantly acting murine allele of Mcm4 causes chromosomal abnormalities and promotes tumorigenesis
Abstract
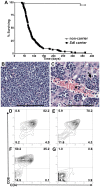
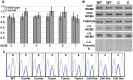
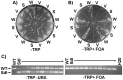
Here we report the isolation of a murine model for heritable T cell lymphoblastic leukemia/lymphoma (T-ALL) called Spontaneous dominant leukemia (Sdl). Sdl heterozygous mice develop disease with a short latency and high penetrance, while mice homozygous for the mutation die early during embryonic development. Sdl mice exhibit an increase in the frequency of micronucleated reticulocytes, and T-ALLs from Sdl mice harbor small amplifications and deletions, including activating deletions at the Notch1 locus. Using exome sequencing it was determined that Sdl mice harbor a spontaneously acquired mutation in Mcm4 (Mcm4(D573H)). MCM4 is part of the heterohexameric complex of MCM2-7 that is important for licensing of DNA origins prior to S phase and also serves as the core of the replicative helicase that unwinds DNA at replication forks. Previous studies in murine models have discovered that genetic reductions of MCM complex levels promote tumor formation by causing genomic instability. However, Sdl mice possess normal levels of Mcms, and there is no evidence for loss-of-heterozygosity at the Mcm4 locus in Sdl leukemias. Studies in Saccharomyces cerevisiae indicate that the Sdl mutation produces a biologically inactive helicase. Together, these data support a model in which chromosomal abnormalities in Sdl mice result from the ability of MCM4(D573H) to incorporate into MCM complexes and render them inactive. Our studies indicate that dominantly acting alleles of MCMs can be compatible with viability but have dramatic oncogenic consequences by causing chromosomal abnormalities.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Shima N, Alcaraz A, Liachko I, Buske TR, Andrews CA, et al. (2007) A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nature genetics 39: 93–98. - PubMed
-
- Pruitt SC, Bailey KJ, Freeland A (2007) Reduced Mcm2 expression results in severe stem/progenitor cell deficiency and cancer. Stem Cells 25: 3121–3132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

