Molecular cloning, expression and molecular modeling of chemosensory protein from Spodoptera litura and its binding properties with Rhodojaponin III
- PMID: 23133516
- PMCID: PMC3485014
- DOI: 10.1371/journal.pone.0047611
Molecular cloning, expression and molecular modeling of chemosensory protein from Spodoptera litura and its binding properties with Rhodojaponin III
Abstract
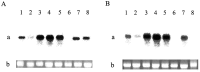
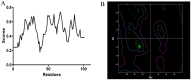
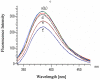
Insects stimulate specific behaviors by the correct recognition of the chemicals in the external environment. Rhodojaponin III is a botanical grayanoid diterpenid oviposition deterrent isolated from Rhododendron molle. In this study we aimed to determine whether the CSPs involved in the recognition of Rhodojaponin III. A full-length cDNA encoding chemosensory protein was isolated from the antennae of Spodoptera litura Fabricius (CSPSlit, GenBank Accession No. DQ007458). The full-length cDNA of NlFoxA is 1789 bp and has an open reading frame (ORF) of 473 bp, encoding a protein of 126 amino acids, Northern blot analysis revealed that CSPSlit mRNA was mainly expressed in the antennae, legs, wings and female abdomens. A three-dimensional model of CSPSlit was constructed using homology modeling method, and its reliability was evaluated. The active site of CSPSlit was calculated using CDOCKER program indicated that the Tyr24, Ile45, Leu49, Thr64, Leu68, Trp79 and Leu82 were responsible ligand-binding active site on identifying Rhodojaponin III in the CSPSlit. The recombinant CSPSlit protein was expressed in Escherichia coli and purified using single-step Ni-NTA affinity chromatography. Fluorescence emission spectra revealed that the CSPSlit protein had significant affinity to rhodojaponin III. These results mean that CSPSlit is critical for insects identify the Rhodojaponin III.
Conflict of interest statement
Figures





References
-
- Awmack CS, Leather SR (2002) Host plant quality and fecundity in herbivorous insects. Annu Rev Entomol 47: 817–844. - PubMed
-
- Hallem EA, Dahanukar A, Carlson JR (2006) Insect odor and taste receptors. Annu Rev Entomol 51: 113–135. - PubMed
-
- Simmonds MS (2001) Importance of flavonoids in insect–plant interactions: feeding and oviposition. Phytochemistry 56: 245–252. - PubMed
-
- Tsuchihara K, Ueno K, Yamanaka A, Isono K, Endo K, et al. (2000) A putative binding protein for lipophilic substances related to butterfly oviposition. FEBS Lett 478: 299–303. - PubMed
-
- Nishida R, Fukami H (1989) Oviposition stimulants of an Aristolochiaceae-feeding swallowtail butterfly, Atrophaneura alcinous . J Chem Ecol 15: 2565–2575. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

