Modulation of the osteosarcoma expression phenotype by microRNAs
- PMID: 23133552
- PMCID: PMC3485010
- DOI: 10.1371/journal.pone.0048086
Modulation of the osteosarcoma expression phenotype by microRNAs
Abstract
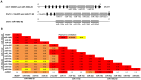
Background: Osteosarcomas are the most common primary malignant tumors of bone and show multiple and complex genomic aberrations. miRNAs are non-coding RNAs capable of regulating gene expression at the post transcriptional level, and miRNAs and their target genes may represent novel therapeutic targets or biomarkers for osteosarcoma. In order to investigate the involvement of miRNAs in osteosarcoma development, global microarray analyses of a panel of 19 human osteosarcoma cell lines was performed.
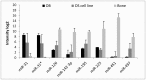
Principal findings: We identified 177 miRNAs that were differentially expressed in osteosarcoma cell lines relative to normal bone. Among these, miR-126/miR-126*, miR-142-3p, miR-150, miR-223, miR-486-5p and members of the miR-1/miR-133a, miR-144/miR-451, miR-195/miR-497 and miR-206/miR-133b clusters were found to be downregulated in osteosarcoma cell lines. All miRNAs in the paralogous clusters miR-17-92, miR-106b-25 and miR-106a-92 were overexpressed. Furthermore, the upregulated miRNAs included miR-9/miR-9*, miR-21*, miR-31/miR-31*, miR-196a/miR-196b, miR-374a and members of the miR-29 and miR-130/301 families. The most interesting inversely correlated miRNA/mRNA pairs in osteosarcoma cell lines included miR-9/TGFBR2 and miR-29/p85α regulatory subunit of PI3K. PTEN mRNA correlated inversely with miR-92a and members of the miR-17 and miR-130/301 families. Expression profiles of selected miRNAs were confirmed in clinical samples. A set of miRNAs, miR-1, miR-18a, miR-18b, miR-19b, miR-31, miR-126, miR-142-3p, miR-133b, miR-144, miR-195, miR-223, miR-451 and miR-497 was identified with an intermediate expression level in osteosarcoma clinical samples compared to osteoblasts and bone, which may reflect the differentiation level of osteosarcoma relative to the undifferentiated osteoblast and fully differentiated normal bone.
Significance: This study provides an integrated analysis of miRNA and mRNA in osteosarcoma, and gives new insight into the complex genetic mechanisms of osteosarcoma development and progression.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

