In vitro site selection of a consensus binding site for the Drosophila melanogaster Tbx20 homolog midline
- PMID: 23133562
- PMCID: PMC3485041
- DOI: 10.1371/journal.pone.0048176
In vitro site selection of a consensus binding site for the Drosophila melanogaster Tbx20 homolog midline
Abstract
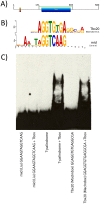
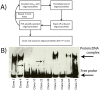
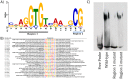
We employed in vitro site selection to identify a consensus binding sequence for the Drosophila melanogaster Tbx20 T-box transcription factor homolog Midline. We purified a bacterially expressed T-box DNA binding domain of Midline, and used it in four rounds of precipitation and polymerase-chain-reaction based amplification. We cloned and sequenced 54 random oligonucleotides selected by Midline. Electromobility shift-assays confirmed that 27 of these could bind the Midline T-box. Sequence alignment of these 27 clones suggests that Midline binds as a monomer to a consensus sequence that contains an AGGTGT core. Thus, the Midline consensus binding site we define in this study is similar to that defined for vertebrate Tbx20, but differs from a previously reported Midline binding sequence derived through site selection.
Conflict of interest statement
Figures

Similar articles
-
The Tbx20 homolog Midline represses wingless in conjunction with Groucho during the maintenance of segment polarity.Dev Biol. 2012 Sep 15;369(2):319-29. doi: 10.1016/j.ydbio.2012.07.004. Epub 2012 Jul 16. Dev Biol. 2012. PMID: 22814213
-
The Tbx20 homologs midline and H15 specify ventral fate in the Drosophila melanogaster leg.Development. 2009 Aug;136(16):2689-93. doi: 10.1242/dev.037911. Epub 2009 Jul 15. Development. 2009. PMID: 19605497
-
Tinman is a direct activator of midline in the Drosophila dorsal vessel.Dev Dyn. 2011 Jan;240(1):86-95. doi: 10.1002/dvdy.22495. Dev Dyn. 2011. PMID: 21108319
-
T-Box Genes in Drosophila Mesoderm Development.Curr Top Dev Biol. 2017;122:161-193. doi: 10.1016/bs.ctdb.2016.06.003. Epub 2016 Jul 30. Curr Top Dev Biol. 2017. PMID: 28057263 Review.
-
T-Box Genes in Drosophila Limb Development.Curr Top Dev Biol. 2017;122:313-354. doi: 10.1016/bs.ctdb.2016.08.003. Epub 2016 Sep 9. Curr Top Dev Biol. 2017. PMID: 28057269 Review.
Cited by
-
Dynamics of promoter bivalency and RNAP II pausing in mouse stem and differentiated cells.BMC Dev Biol. 2018 Feb 20;18(1):2. doi: 10.1186/s12861-018-0163-7. BMC Dev Biol. 2018. PMID: 29458328 Free PMC article.
-
midline represses Dpp signaling and target gene expression in Drosophila ventral leg development.Biol Open. 2022 May 15;11(5):bio059206. doi: 10.1242/bio.059206. Epub 2022 May 24. Biol Open. 2022. PMID: 35608103 Free PMC article.
-
The expression of the T-box selector gene midline in the leg imaginal disc is controlled by both transcriptional regulation and cell lineage.Biol Open. 2015 Nov 18;4(12):1707-14. doi: 10.1242/bio.013565. Biol Open. 2015. PMID: 26581591 Free PMC article.
References
-
- Papaioannou VE (2001) T-box genes in development: from hydra to humans. Int Rev Cytol 207: 1–70. - PubMed
-
- Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE (2005) T-box genes in vertebrate development. Annu Rev Genet 39: 219–239. - PubMed
-
- Ghosh TK, Packham EA, Bonser AJ, Robinson TE, Cross SJ, et al. (2001) Characterization of the TBX5 binding site and analysis of mutations that cause Holt-Oram syndrome. Hum Mol Genet 10: 1983–1994. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

