Genomic comparison of Escherichia coli O104:H4 isolates from 2009 and 2011 reveals plasmid, and prophage heterogeneity, including shiga toxin encoding phage stx2
- PMID: 23133618
- PMCID: PMC3486847
- DOI: 10.1371/journal.pone.0048228
Genomic comparison of Escherichia coli O104:H4 isolates from 2009 and 2011 reveals plasmid, and prophage heterogeneity, including shiga toxin encoding phage stx2
Abstract
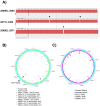
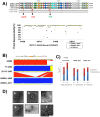
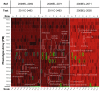
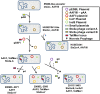
In May of 2011, an enteroaggregative Escherichia coli O104:H4 strain that had acquired a Shiga toxin 2-converting phage caused a large outbreak of bloody diarrhea in Europe which was notable for its high prevalence of hemolytic uremic syndrome cases. Several studies have described the genomic inventory and phylogenies of strains associated with the outbreak and a collection of historical E. coli O104:H4 isolates using draft genome assemblies. We present the complete, closed genome sequences of an isolate from the 2011 outbreak (2011C-3493) and two isolates from cases of bloody diarrhea that occurred in the Republic of Georgia in 2009 (2009EL-2050 and 2009EL-2071). Comparative genome analysis indicates that, while the Georgian strains are the nearest neighbors to the 2011 outbreak isolates sequenced to date, structural and nucleotide-level differences are evident in the Stx2 phage genomes, the mer/tet antibiotic resistance island, and in the prophage and plasmid profiles of the strains, including a previously undescribed plasmid with homology to the pMT virulence plasmid of Yersinia pestis. In addition, multiphenotype analysis showed that 2009EL-2071 possessed higher resistance to polymyxin and membrane-disrupting agents. Finally, we show evidence by electron microscopy of the presence of a common phage morphotype among the European and Georgian strains and a second phage morphotype among the Georgian strains. The presence of at least two stx2 phage genotypes in host genetic backgrounds that may derive from a recent common ancestor of the 2011 outbreak isolates indicates that the emergence of stx2 phage-containing E. coli O104:H4 strains probably occurred more than once, or that the current outbreak isolates may be the result of a recent transfer of a new stx2 phage element into a pre-existing stx2-positive genetic background.
Conflict of interest statement
Figures




References
-
- Kaper JB, Nataro JP, Mobley HL (2004) Pathogenic Escherichia coli. Nat Rev Microbiol 2: 123–140. - PubMed
-
- DuPont HL (2007) The growing threat of foodborne bacterial enteropathogens of animal origin. Clin Infect Dis 45: 1353–1361. - PubMed
-
- Barlow RS, Gobius KS, Desmarchelier PM (2006) Shiga toxin-producing Escherichia coli in ground beef and lamb cuts: results of a one-year study. Int J Food Microbiol 111: 1–5. - PubMed
-
- Berger CN, Sodha SV, Shaw RK, Griffin PM, Pink D, et al. (2010) Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environ Microbiol 12: 2385–2397. - PubMed
-
- Brandl MT (2006) Fitness of human enteric pathogens on plants and implications for food safety. Annu Rev Phytopathol 44: 367–392. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

