Distribution and quantification of antibiotic resistant genes and bacteria across agricultural and non-agricultural metagenomes
- PMID: 23133629
- PMCID: PMC3487761
- DOI: 10.1371/journal.pone.0048325
Distribution and quantification of antibiotic resistant genes and bacteria across agricultural and non-agricultural metagenomes
Abstract
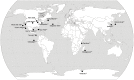
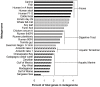
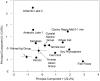
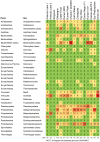
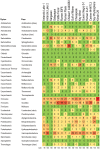
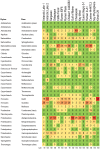
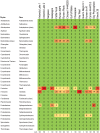
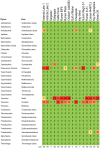
There is concern that antibiotic resistance can potentially be transferred from animals to humans through the food chain. The relationship between specific antibiotic resistant bacteria and the genes they carry remains to be described. Few details are known about the ecology of antibiotic resistant genes and bacteria in food production systems, or how antibiotic resistance genes in food animals compare to antibiotic resistance genes in other ecosystems. Here we report the distribution of antibiotic resistant genes in publicly available agricultural and non-agricultural metagenomic samples and identify which bacteria are likely to be carrying those genes. Antibiotic resistance, as coded for in the genes used in this study, is a process that was associated with all natural, agricultural, and human-impacted ecosystems examined, with between 0.7 to 4.4% of all classified genes in each habitat coding for resistance to antibiotic and toxic compounds (RATC). Agricultural, human, and coastal-marine metagenomes have characteristic distributions of antibiotic resistance genes, and different bacteria that carry the genes. There is a larger percentage of the total genome associated with antibiotic resistance in gastrointestinal-associated and agricultural metagenomes compared to marine and Antarctic samples. Since antibiotic resistance genes are a natural part of both human-impacted and pristine habitats, presence of these resistance genes in any specific habitat is therefore not sufficient to indicate or determine impact of anthropogenic antibiotic use. We recommend that baseline studies and control samples be taken in order to determine natural background levels of antibiotic resistant bacteria and/or antibiotic resistance genes when investigating the impacts of veterinary use of antibiotics on human health. We raise questions regarding whether the underlying biology of each type of bacteria contributes to the likelihood of transfer via the food chain.
Conflict of interest statement
Figures
Similar articles
-
Metagenomic investigation reveals bacteriophage-mediated horizontal transfer of antibiotic resistance genes in microbial communities of an organic agricultural ecosystem.Microbiol Spectr. 2023 Oct 17;11(5):e0022623. doi: 10.1128/spectrum.00226-23. Epub 2023 Sep 27. Microbiol Spectr. 2023. PMID: 37754684 Free PMC article.
-
Metagenomic analysis of virulence-associated and antibiotic resistance genes of microbes in rumen of Indian buffalo (Bubalus bubalis).Gene. 2012 Oct 10;507(2):146-51. doi: 10.1016/j.gene.2012.07.037. Epub 2012 Jul 28. Gene. 2012. PMID: 22850272
-
Antibiotic resistomes of healthy pig faecal metagenomes.Microb Genom. 2019 May;5(5):e000272. doi: 10.1099/mgen.0.000272. Epub 2019 May 15. Microb Genom. 2019. PMID: 31091181 Free PMC article.
-
Tetracycline and Phenicol Resistance Genes and Mechanisms: Importance for Agriculture, the Environment, and Humans.J Environ Qual. 2016 Mar;45(2):576-92. doi: 10.2134/jeq2015.04.0207. J Environ Qual. 2016. PMID: 27065405 Review.
-
The incidence of antibiotic resistance within and beyond the agricultural ecosystem: A concern for public health.Microbiologyopen. 2020 Sep;9(9):e1035. doi: 10.1002/mbo3.1035. Epub 2020 Jul 25. Microbiologyopen. 2020. PMID: 32710495 Free PMC article. Review.
Cited by
-
Diversity of bacterial communities in wetlands of Calakmul Biosphere Reserve: a comparative analysis between conserved and semi-urbanized zones in pre-Mayan Train era.BMC Microbiol. 2024 Sep 28;24(1):376. doi: 10.1186/s12866-024-03523-x. BMC Microbiol. 2024. PMID: 39342129 Free PMC article.
-
Characterization of antibiogram fingerprints in Listeria monocytogenes recovered from irrigation water and agricultural soil samples.PLoS One. 2020 Feb 10;15(2):e0228956. doi: 10.1371/journal.pone.0228956. eCollection 2020. PLoS One. 2020. PMID: 32040533 Free PMC article.
-
Recovery of Previously Uncultured Bacterial Genera from Three Mediterranean Sponges.Mar Biotechnol (NY). 2017 Oct;19(5):454-468. doi: 10.1007/s10126-017-9766-4. Epub 2017 Jul 10. Mar Biotechnol (NY). 2017. PMID: 28695385 Free PMC article.
-
Agriculturally Sourced Multidrug-Resistant Escherichia coli for Use as Control Strains.Pathogens. 2025 Apr 25;14(5):417. doi: 10.3390/pathogens14050417. Pathogens. 2025. PMID: 40430738 Free PMC article.
-
High genomic diversity of multi-drug resistant wastewater Escherichia coli.Sci Rep. 2018 Jun 12;8(1):8928. doi: 10.1038/s41598-018-27292-6. Sci Rep. 2018. PMID: 29895899 Free PMC article.
References
-
- Collignon P, Powers JH, Chiller TM, Aidara-Kane A, Aarestrup AFM (2009) World Health Organization ranking of antimicrobials according to their importance in human medicine: a critical step for developing risk management strategies for the use of antimicrobials in food production animals. Clin Infect Dis 49: 132–141. - PubMed
-
- Crippen TL, Poole TL (2009) Conjugative transfer of plasmid-located antibiotic resistance genes within the gastrointestinal tract of lesser mealworm larvae, Alphitobius diaperinius (Coleoptera: Tenebrionidae). Foodborne Pathog Dis 7: 907–915. - PubMed
-
- Teuber M (2001) Veterinary use and antibiotic resistance. Curr Opin Microbiol 4: 493–499. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

