Revealing the hyperdiverse mite fauna of subarctic Canada through DNA barcoding
- PMID: 23133656
- PMCID: PMC3487733
- DOI: 10.1371/journal.pone.0048755
Revealing the hyperdiverse mite fauna of subarctic Canada through DNA barcoding
Abstract
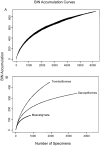
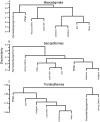
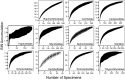
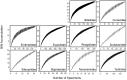
Although mites are one of the most abundant and diverse groups of arthropods, they are rarely targeted for detailed biodiversity surveys due to taxonomic constraints. We address this gap through DNA barcoding, evaluating acarine diversity at Churchill, Manitoba, a site on the tundra-taiga transition. Barcode analysis of 6279 specimens revealed nearly 900 presumptive species of mites with high species turnover between substrates and between forested and non-forested sites. Accumulation curves have not reached an asymptote for any of the three mite orders investigated, and estimates suggest that more than 1200 species of Acari occur at this locality. The coupling of DNA barcode results with taxonomic assignments revealed that Trombidiformes compose 49% of the fauna, a larger fraction than expected based on prior studies. This investigation demonstrates the efficacy of DNA barcoding in facilitating biodiversity assessments of hyperdiverse taxa.
Conflict of interest statement
Figures



Similar articles
-
Spiders (Araneae) of Churchill, Manitoba: DNA barcodes and morphology reveal high species diversity and new Canadian records.BMC Ecol. 2013 Nov 26;13:44. doi: 10.1186/1472-6785-13-44. BMC Ecol. 2013. PMID: 24279427 Free PMC article.
-
DNA barcodes expose unexpected diversity in Canadian mites.Mol Ecol. 2019 Dec;28(24):5347-5359. doi: 10.1111/mec.15292. Epub 2019 Nov 22. Mol Ecol. 2019. PMID: 31674085
-
DNA barcoding for community ecology--how to tackle a hyperdiverse, mostly undescribed Melanesian fauna.PLoS One. 2012;7(1):e28832. doi: 10.1371/journal.pone.0028832. Epub 2012 Jan 13. PLoS One. 2012. PMID: 22253699 Free PMC article.
-
The importance of taxonomic resolution for additive beta diversity as revealed through DNA barcoding.Genome. 2016 Dec;59(12):1130-1140. doi: 10.1139/gen-2016-0080. Epub 2016 Aug 18. Genome. 2016. PMID: 27845571 Review.
-
Molecular-based rapid inventories of sympatric diversity: a comparison of DNA barcode clustering methods applied to geography-based vs clade-based sampling of amphibians.J Biosci. 2012 Nov;37(5):887-96. doi: 10.1007/s12038-012-9255-x. J Biosci. 2012. PMID: 23107924 Review.
Cited by
-
Species Identification in Malaise Trap Samples by DNA Barcoding Based on NGS Technologies and a Scoring Matrix.PLoS One. 2016 May 18;11(5):e0155497. doi: 10.1371/journal.pone.0155497. eCollection 2016. PLoS One. 2016. PMID: 27191722 Free PMC article.
-
The genus Vipio Latreille (Hymenoptera, Braconidae) in the Neotropical Region.Zookeys. 2020 Apr 8;925:89-140. doi: 10.3897/zookeys.925.48457. eCollection 2020. Zookeys. 2020. PMID: 32390742 Free PMC article.
-
Biodiversity inventories in high gear: DNA barcoding facilitates a rapid biotic survey of a temperate nature reserve.Biodivers Data J. 2015 Aug 30;(3):e6313. doi: 10.3897/BDJ.3.e6313. eCollection 2015. Biodivers Data J. 2015. PMID: 26379469 Free PMC article.
-
Towards a Canary Islands barcode database for soil biodiversity: revealing cryptic and unrecorded mite species diversity within insular soils.Biodivers Data J. 2024 Jan 10;12:e113301. doi: 10.3897/BDJ.12.e113301. eCollection 2024. Biodivers Data J. 2024. PMID: 38314123 Free PMC article.
-
Phylogeny and species delineation in European species of the genus Steganacarus (Acari, Oribatida) using mitochondrial and nuclear markers.Exp Appl Acarol. 2015 Jun;66(2):173-86. doi: 10.1007/s10493-015-9905-4. Epub 2015 Apr 10. Exp Appl Acarol. 2015. PMID: 25860859
References
-
- Zhou X, Adamowicz SJ, Jacobus LM, DeWalt RE, Hebert PDN (2009) Towards a comprehensive barcode library for arctic life – Ephemeroptera, Plecoptera, and Trichoptera of Churchill, Manitoba, Canada. Frontiers in Zoology 6: 30 doi:10.1186/1742-9994-6-30. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

