Phylogeny of the genus Chrysanthemum L.: evidence from single-copy nuclear gene and chloroplast DNA sequences
- PMID: 23133665
- PMCID: PMC3486802
- DOI: 10.1371/journal.pone.0048970
Phylogeny of the genus Chrysanthemum L.: evidence from single-copy nuclear gene and chloroplast DNA sequences
Abstract
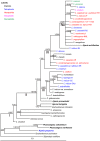
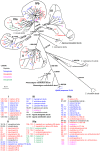
Chrysanthemum L. (Asteraceae-Anthemideae) is a genus with rapid speciation. It comprises about 40 species, most of which are distributed in East Asia. Many of these are narrowly distributed and habitat-specific. Considerable variations in morphology and ploidy are found in this genus. Some species have been the subjects of many studies, but the relationships between Chrysanthemum and its allies and the phylogeny of this genus remain poorly understood. In the present study, 32 species/varieties from Chrysanthemum and 11 from the allied genera were analyzed using DNA sequences of the single-copy nuclear CDS gene and seven cpDNA loci (psbA-trnH, trnC-ycf6, ycf6-psbM, trnY-rpoB, rpS4-trnT, trnL-F, and rpL16). The cpDNA and nuclear CDS gene trees both suggest that 1) Chrysanthemum is not a monophyletic taxon, and the affinity between Chrysanthemum and Ajania is so close that these two genera should be incorporated taxonomically; 2) Phaeostigma is more closely related to the Chrysanthemum+Ajania than other generic allies. According to pollen morphology and to the present cpDNA and CDS data, Ajania purpurea is a member of Phaeostigma. Species differentiation in Chrysanthemum appears to be correlated with geographic and environmental conditions. The Chinese Chrysanthemum species can be divided into two groups, the C. zawadskii group and the C. indicum group. The former is distributed in northern China and the latter in southern China. Many polyploid species, such as C. argyrophyllum, may have originated from allopolyploidization involving divergent progenitors. Considering all the evidence from present and previous studies, we conclude that geographic and ecological factors as well as hybridization and polyploidy play important roles in the divergence and speciation of the genus Chrysanthemum.
Conflict of interest statement
Figures
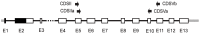


References
-
- Kondo K, Abd El-Twab MH, Idesawa R, Kimura S, Tanaka R (2003) Genome phylogenetics in Chrysanthemum sensu lato. In: Sharma AK, Sharma A, editors. Plant Genome: Biodiversity and Evolution, Vol 1A, Phanerogams. Plymouth: Science Publisher. 117–200.
-
- Yang WH, Glover BJ, Rao GY, Yang J (2006) Molecular evidence for multiple polyploidization and lineage recombination in the Chrysanthemum indicum polyploid complex (Asteraceae). New Phytol 171: 875–886. - PubMed
-
- Nataka M, Tanaka R (1987) Species of Chrysanthemum in Japan in the study of chromosomes. In: Hong DY, editor. Plant Chromsome Research. Hiroshima, Japan: Nishiki. 17–22.
-
- Tsukaya H (2002) Leaf anatomy of a rheophyte, Dendranthema yoshinaganthum (Asteraceae), and of hybrids between D. yoshinaganthum and a closely related non-rheophyte, D. indicum . J Plant Res 115: 329–333. - PubMed
-
- Kim JS, Pak JH, Seo BB, Tobe H (2003) Karyotypes of metaphase chromosomes in diploid populations of Dendranthema zawadskii and related species (Asteraceae) from Korea: diversity and evolutionary implications. J Plant Res 116: 47–55. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

