Extension of yeast chronological lifespan by methylamine
- PMID: 23133668
- PMCID: PMC3487785
- DOI: 10.1371/journal.pone.0048982
Extension of yeast chronological lifespan by methylamine
Abstract
Background: Chronological aging of yeast cells is commonly used as a model for aging of human post-mitotic cells. The yeast Saccharomyces cerevisiae grown on glucose in the presence of ammonium sulphate is mainly used in yeast aging research. We have analyzed chronological aging of the yeast Hansenula polymorpha grown at conditions that require primary peroxisome metabolism for growth.
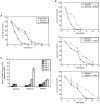
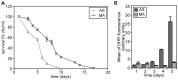
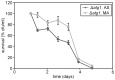
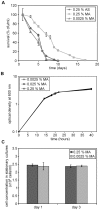
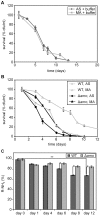
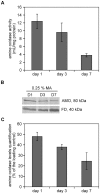
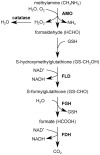
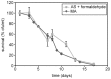
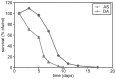
Methodology/principal findings: The chronological lifespan of H. polymorpha is strongly enhanced when cells are grown on methanol or ethanol, metabolized by peroxisome enzymes, relative to growth on glucose that does not require peroxisomes. The short lifespan of H. polymorpha on glucose is mainly due to medium acidification, whereas most likely ROS do not play an important role. Growth of cells on methanol/methylamine instead of methanol/ammonium sulphate resulted in further lifespan enhancement. This was unrelated to medium acidification. We show that oxidation of methylamine by peroxisomal amine oxidase at carbon starvation conditions is responsible for lifespan extension. The methylamine oxidation product formaldehyde is further oxidized resulting in NADH generation, which contributes to increased ATP generation and reduction of ROS levels in the stationary phase.
Conclusion/significance: We conclude that primary peroxisome metabolism enhanced chronological lifespan of H. polymorpha. Moreover, the possibility to generate NADH at carbon starvation conditions by an organic nitrogen source supports further extension of the lifespan of the cell. Consequently, the interpretation of CLS analyses in yeast should include possible effects on the energy status of the cell.
Conflict of interest statement
Figures
References
-
- Kennedy BK (2008) The genetics of ageing: insight from genome-wide approaches in invertebrate model organisms. J Intern Med 263: 142–152. - PubMed
-
- MacLean M, Harris N, Piper PW (2001) Chronological lifespan of stationary phase yeast cells; a model for investigating the factors that might influence the ageing of postmitotic tissues in higher organisms. Yeast 18: 499–509. - PubMed
-
- Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD (2001) Regulation of longevity and stress resistance by Sch9 in yeast. Science 292: 288–290. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

