Epigenetics-beyond the genome in alcoholism
- PMID: 23134045
- PMCID: PMC3860414
Epigenetics-beyond the genome in alcoholism
Abstract
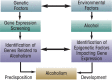
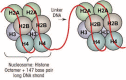
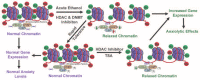
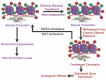
Genetic and environmental factors play a role in the development of alcoholism. Whole-genome expression profiling has highlighted the importance of several genes that may contribute to alcohol abuse disorders. In addition, more recent findings have added yet another layer of complexity to the overall molecular mechanisms involved in a predisposition to alcoholism and addiction by demonstrating that processes related to genetic factors that do not manifest as DNA sequence changes (i.e., epigenetic processes) play a role. Both acute and chronic ethanol exposure can alter gene expression levels in specific neuronal circuits that govern the behavioral consequences related to tolerance and dependence. The unremitting cycle of alcohol consumption often includes satiation and self-medication with alcohol, followed by excruciating withdrawal symptoms and the resultant relapse, which reflects both the positive and negative affective states of alcohol addiction. Recent studies have indicated that behavioral changes induced by acute and chronic ethanol exposure may involve chromatin remodeling resulting from covalent histone modifications and DNA methylation in the neuronal circuits involving a brain region called the amygdala. These findings have helped identify enzymes involved in epigenetic mechanisms, such as the histone deacetylase, histone acetyltransferase, and DNA methyltransferase enzymes, as novel therapeutic targets for the development of future pharmacotherapies for the treatment of alcoholism.
Figures
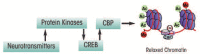
References
-
- Agger K, Christensen J, Cloos PA, Helin K. The emerging functions of histone demethylases. Current Opinion in Genetics and Development. 2008;18:159–168. - PubMed
-
- Alarcon JM, Malleret G, Touzani K, et al. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: A model for the cognitive deficit in Rubenstein-Taybi Syndrome and its amelioration. Neuron. 2004;42:947–959. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

