Effects of multiple conformers per compound upon 3-D similarity search and bioassay data analysis
- PMID: 23134593
- PMCID: PMC3537644
- DOI: 10.1186/1758-2946-4-28
Effects of multiple conformers per compound upon 3-D similarity search and bioassay data analysis
Abstract
Background: To improve the utility of PubChem, a public repository containing biological activities of small molecules, the PubChem3D project adds computationally-derived three-dimensional (3-D) descriptions to the small-molecule records contained in the PubChem Compound database and provides various search and analysis tools that exploit 3-D molecular similarity. Therefore, the efficient use of PubChem3D resources requires an understanding of the statistical and biological meaning of computed 3-D molecular similarity scores between molecules.
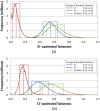
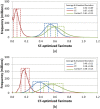
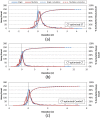
Results: The present study investigated effects of employing multiple conformers per compound upon the 3-D similarity scores between ten thousand randomly selected biologically-tested compounds (10-K set) and between non-inactive compounds in a given biological assay (156-K set). When the "best-conformer-pair" approach, in which a 3-D similarity score between two compounds is represented by the greatest similarity score among all possible conformer pairs arising from a compound pair, was employed with ten diverse conformers per compound, the average 3-D similarity scores for the 10-K set increased by 0.11, 0.09, 0.15, 0.16, 0.07, and 0.18 for STST-opt, CTST-opt, ComboTST-opt, STCT-opt, CTCT-opt, and ComboTCT-opt, respectively, relative to the corresponding averages computed using a single conformer per compound. Interestingly, the best-conformer-pair approach also increased the average 3-D similarity scores for the non-inactive-non-inactive (NN) pairs for a given assay, by comparable amounts to those for the random compound pairs, although some assays showed a pronounced increase in the per-assay NN-pair 3-D similarity scores, compared to the average increase for the random compound pairs.
Conclusion: These results suggest that the use of ten diverse conformers per compound in PubChem bioassay data analysis using 3-D molecular similarity is not expected to increase the separation of non-inactive from random and inactive spaces "on average", although some assays show a noticeable separation between the non-inactive and random spaces when multiple conformers are used for each compound. The present study is a critical next step to understand effects of conformational diversity of the molecules upon the 3-D molecular similarity and its application to biological activity data analysis in PubChem. The results of this study may be helpful to build search and analysis tools that exploit 3-D molecular similarity between compounds archived in PubChem and other molecular libraries in a more efficient way.
Figures









Similar articles
-
PubChem3D: Biologically relevant 3-D similarity.J Cheminform. 2011 Jul 22;3(1):26. doi: 10.1186/1758-2946-3-26. J Cheminform. 2011. PMID: 21781288 Free PMC article.
-
PubChem3D: conformer ensemble accuracy.J Cheminform. 2013 Jan 7;5(1):1. doi: 10.1186/1758-2946-5-1. J Cheminform. 2013. PMID: 23289532 Free PMC article.
-
PubChem3D: Similar conformers.J Cheminform. 2011 May 9;3:13. doi: 10.1186/1758-2946-3-13. J Cheminform. 2011. PMID: 21554721 Free PMC article.
-
Chemical Structure Similarity Search for Ligand-based Virtual Screening: Methods and Computational Resources.Curr Drug Targets. 2016;17(14):1580-1585. doi: 10.2174/1389450116666151102095555. Curr Drug Targets. 2016. PMID: 26521773 Review.
-
Benchmarking Data Sets from PubChem BioAssay Data: Current Scenario and Room for Improvement.Int J Mol Sci. 2020 Jun 19;21(12):4380. doi: 10.3390/ijms21124380. Int J Mol Sci. 2020. PMID: 32575564 Free PMC article. Review.
Cited by
-
An update on PUG-REST: RESTful interface for programmatic access to PubChem.Nucleic Acids Res. 2018 Jul 2;46(W1):W563-W570. doi: 10.1093/nar/gky294. Nucleic Acids Res. 2018. PMID: 29718389 Free PMC article.
-
Similar compounds versus similar conformers: complementarity between PubChem 2-D and 3-D neighboring sets.J Cheminform. 2016 Nov 4;8:62. doi: 10.1186/s13321-016-0163-1. eCollection 2016. J Cheminform. 2016. PMID: 27872662 Free PMC article.
-
Integration of mass spectral fingerprinting analysis with precursor ion (MS1) quantification for the characterisation of botanical extracts: application to extracts of Centella asiatica (L.) Urban.Phytochem Anal. 2020 Nov;31(6):722-738. doi: 10.1002/pca.2936. Epub 2020 Apr 12. Phytochem Anal. 2020. PMID: 32281154 Free PMC article.
-
Target enhanced 2D similarity search by using explicit biological activity annotations and profiles.J Cheminform. 2015 Nov 17;7:55. doi: 10.1186/s13321-015-0103-5. eCollection 2015. J Cheminform. 2015. PMID: 26583046 Free PMC article.
-
Finding Potential Multitarget Ligands Using PubChem.Methods Mol Biol. 2018;1825:63-91. doi: 10.1007/978-1-4939-8639-2_2. Methods Mol Biol. 2018. PMID: 30334203 Free PMC article.
References
-
- Bolton EE, Wang Y, Thiessen PA, Bryant SH. In: Annual Reports in Computational Chemistry. Volume 4. Ralph AW, David CS, editor. Amsterdam, the Netherlands: Elsevier; 2008. PubChem: integrated platform of small molecules and biological activities; pp. 217–241.
-
- PubChem3D Thematic Series. http://www.jcheminf.com/series/pubchem3d.
LinkOut - more resources
Full Text Sources
Other Literature Sources

