ST proteins, a new family of plant tandem repeat proteins with a DUF2775 domain mainly found in Fabaceae and Asteraceae
- PMID: 23134664
- PMCID: PMC3499167
- DOI: 10.1186/1471-2229-12-207
ST proteins, a new family of plant tandem repeat proteins with a DUF2775 domain mainly found in Fabaceae and Asteraceae
Abstract
Background: Many proteins with tandem repeats in their sequence have been described and classified according to the length of the repeats: I) Repeats of short oligopeptides (from 2 to 20 amino acids), including structural cell wall proteins and arabinogalactan proteins. II) Repeats that range in length from 20 to 40 residues, including proteins with a well-established three-dimensional structure often involved in mediating protein-protein interactions. (III) Longer repeats in the order of 100 amino acids that constitute structurally and functionally independent units. Here we analyse ShooT specific (ST) proteins, a family of proteins with tandem repeats of unknown function that were first found in Leguminosae, and their possible similarities to other proteins with tandem repeats.
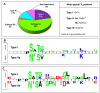
Results: ST protein sequences were only found in dicotyledonous plants, limited to several plant families, mainly the Fabaceae and the Asteraceae. ST mRNAs accumulate mainly in the roots and under biotic interactions. Most ST proteins have one or several Domain(s) of Unknown Function 2775 (DUF2775). All deduced ST proteins have a signal peptide, indicating that these proteins enter the secretory pathway, and the mature proteins have tandem repeat oligopeptides that share a hexapeptide (E/D)FEPRP followed by 4 partially conserved amino acids, which could determine a putative N-glycosylation signal, and a fully conserved tyrosine. In a phylogenetic tree, the sequences clade according to taxonomic group. A possible involvement in symbiosis and abiotic stress as well as in plant cell elongation is suggested, although different STs could play different roles in plant development.
Conclusions: We describe a new family of proteins called ST whose presence is limited to the plant kingdom, specifically to a few families of dicotyledonous plants. They present 20 to 40 amino acid tandem repeat sequences with different characteristics (signal peptide, DUF2775 domain, conservative repeat regions) from the described group of 20 to 40 amino acid tandem repeat proteins and also from known cell wall proteins with repeat sequences. Several putative roles in plant physiology can be inferred from the characteristics found.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

