Identification of a rudimentary neural crest in a non-vertebrate chordate
- PMID: 23135395
- PMCID: PMC4257486
- DOI: 10.1038/nature11589
Identification of a rudimentary neural crest in a non-vertebrate chordate
Abstract
Neural crest arises at the neural plate border, expresses a core set of regulatory genes and produces a diverse array of cell types, including ectomesenchyme derivatives that elaborate the vertebrate head. The evolution of neural crest has been proposed to be a key event leading to the appearance of new cell types that fostered the transition from filter feeding to active predation in ancestral vertebrates. However, the origin of neural crest remains controversial, as homologous cell types have not been unambiguously identified in non-vertebrate chordates. Here we show that the tunicate Ciona intestinalis possesses a cephalic melanocyte lineage (a9.49) similar to neural crest that can be reprogrammed into migrating 'ectomesenchyme' by the targeted misexpression of Twist (also known as twist-like 2). Our results suggest that the neural crest melanocyte regulatory network pre-dated the divergence of tunicates and vertebrates. We propose that the co-option of mesenchyme determinants, such as Twist, into the neural plate ectoderm was crucial to the emergence of the vertebrate 'new head'.
Figures
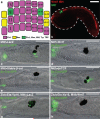
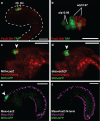
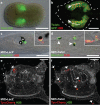
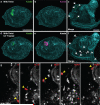
References
-
- Le Douarin NM, et al. Neural crest cell plasticity and its limits. Development. 2004;131:4637–4650. - PubMed
-
- Gans C, Northcutt RG. Neural crest and the origin of vertebrates: a new head. Science. 1983;220:268–273. - PubMed
-
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

