Chromoanagenesis and cancer: mechanisms and consequences of localized, complex chromosomal rearrangements
- PMID: 23135524
- PMCID: PMC3616639
- DOI: 10.1038/nm.2988
Chromoanagenesis and cancer: mechanisms and consequences of localized, complex chromosomal rearrangements
Abstract
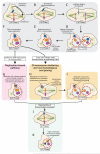
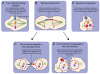
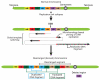
Next-generation sequencing of DNA from human tumors or individuals with developmental abnormalities has led to the discovery of a process we term chromoanagenesis, in which large numbers of complex rearrangements occur at one or a few chromosomal loci in a single catastrophic event. Two mechanisms underlie these rearrangements, both of which can be facilitated by a mitotic chromosome segregation error to produce a micronucleus containing the chromosome to undergo rearrangement. In the first, chromosome shattering (chromothripsis) is produced by mitotic entry before completion of DNA replication within the micronucleus, with a failure to disassemble the micronuclear envelope encapsulating the chromosomal fragments for random reassembly in the subsequent interphase. Alternatively, locally defective DNA replication initiates serial, microhomology-mediated template switching (chromoanasynthesis) that produces local rearrangements with altered gene copy numbers. Complex rearrangements are present in a broad spectrum of tumors and in individuals with congenital or developmental defects, highlighting the impact of chromoanagenesis on human disease.
Figures

Similar articles
-
Scrambling the genome in cancer: causes and consequences of complex chromosome rearrangements.Nat Rev Genet. 2024 Mar;25(3):196-210. doi: 10.1038/s41576-023-00663-0. Epub 2023 Nov 8. Nat Rev Genet. 2024. PMID: 37938738 Free PMC article. Review.
-
Catastrophic cellular events leading to complex chromosomal rearrangements in the germline.Clin Genet. 2017 May;91(5):653-660. doi: 10.1111/cge.12928. Epub 2017 Feb 22. Clin Genet. 2017. PMID: 27888607 Review.
-
Processes shaping cancer genomes - From mitotic defects to chromosomal rearrangements.DNA Repair (Amst). 2021 Nov;107:103207. doi: 10.1016/j.dnarep.2021.103207. Epub 2021 Aug 10. DNA Repair (Amst). 2021. PMID: 34425515 Review.
-
Chromoanagenesis: cataclysms behind complex chromosomal rearrangements.Mol Cytogenet. 2019 Feb 11;12:6. doi: 10.1186/s13039-019-0415-7. eCollection 2019. Mol Cytogenet. 2019. PMID: 30805029 Free PMC article. Review.
-
Chromoanagenesis, the mechanisms of a genomic chaos.Semin Cell Dev Biol. 2022 Mar;123:90-99. doi: 10.1016/j.semcdb.2021.01.004. Epub 2021 Feb 16. Semin Cell Dev Biol. 2022. PMID: 33608210 Review.
Cited by
-
Griseofulvin Radiosensitizes Non-Small Cell Lung Cancer Cells and Activates cGAS.Mol Cancer Ther. 2023 Apr 3;22(4):519-528. doi: 10.1158/1535-7163.MCT-22-0191. Mol Cancer Ther. 2023. PMID: 36752776 Free PMC article.
-
Challenges in the Use of Targeted Therapies in Non-Small Cell Lung Cancer.Cancer Res Treat. 2022 Apr;54(2):315-329. doi: 10.4143/crt.2022.078. Epub 2022 Feb 18. Cancer Res Treat. 2022. PMID: 35209703 Free PMC article. Review.
-
Genome Chaos, Information Creation, and Cancer Emergence: Searching for New Frameworks on the 50th Anniversary of the "War on Cancer".Genes (Basel). 2021 Dec 31;13(1):101. doi: 10.3390/genes13010101. Genes (Basel). 2021. PMID: 35052441 Free PMC article. Review.
-
[Molecular characterization of osteosarcomas].Pathologe. 2013 Nov;34 Suppl 2:260-3. doi: 10.1007/s00292-013-1823-9. Pathologe. 2013. PMID: 24196625 Review. German.
-
Chromothripsis and beyond: rapid genome evolution from complex chromosomal rearrangements.Genes Dev. 2013 Dec 1;27(23):2513-30. doi: 10.1101/gad.229559.113. Genes Dev. 2013. PMID: 24298051 Free PMC article. Review.
References
-
- Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

