Inferring the inter-host transmission of influenza A virus using patterns of intra-host genetic variation
- PMID: 23135678
- PMCID: PMC3574438
- DOI: 10.1098/rspb.2012.2173
Inferring the inter-host transmission of influenza A virus using patterns of intra-host genetic variation
Abstract
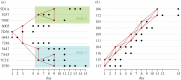
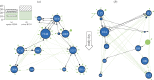
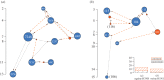
Influenza A viruses (IAVs) cause acute, highly transmissible infections in a wide range of animal species. Understanding how these viruses are transmitted within and between susceptible host populations is critical to the development of effective control strategies. While viral gene sequences have been used to make inferences about IAV transmission dynamics at the epidemiological scale, their utility in accurately determining patterns of inter-host transmission in the short-term--i.e. who infected whom--has not been strongly established. Herein, we use intra-host sequence data from the viral HA1 (hemagglutinin) gene domain from two transmission studies employing different IAV subtypes in their natural hosts--H3N8 in horses and H1N1 in pigs-to determine how well these data recapitulate the known pattern of inter-host transmission. Although no mutations were fixed over the course of either experimental transmission chain, we show that some minor, transient alleles can provide evidence of host-to-host transmission and, importantly, can be distinguished from those that cannot.
Figures
References
-
- Chen R, Holmes EC. 2006. Avian influenza virus exhibits rapid evolutionary dynamics. Mol. Biol. Evol. 23, 2336–234110.1093/molbev/msl102 (doi:10.1093/molbev/msl102) - DOI - DOI - PubMed
-
- Murcia PR, et al. 2010. Intra- and interhost evolutionary dynamics of equine influenza virus. J. Virol. 84, 6943–695410.1128/JVI.00112-10 (doi:10.1128/JVI.00112-10) - DOI - DOI - PMC - PubMed
-
- Ault A, et al. 2012. Immunogenicity and clinical protection against equine influenza by DNA vaccination of ponies. Vaccine 30, 3965–397410.1016/j.vaccine.2012.03.026 (doi:10.1016/j.vaccine.2012.03.026) - DOI - DOI - PMC - PubMed
-
- Lloyd LE, et al. 2011. Experimental transmission of avian-like swine H1N1 influenza virus between immunologically naïve and vaccinated pigs. Influenza Other Respir. Viruses 5, 357–36410.1111/j.1750-2659.2011.00233.x (doi:10.1111/j.1750-2659.2011.00233.x) - DOI - DOI - PMC - PubMed
-
- McHardy AC, Adams B. 2009. The role of genomics in tracking the evolution of influenza A virus. PLoS Pathog. 5, e1000566.10.1371/journal.ppat.1000566 (doi:10.1371/journal.ppat.1000566) - DOI - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

