An integrated genome-wide approach to discover tumor-specific antigens as potential immunologic and clinical targets in cancer
- PMID: 23135912
- PMCID: PMC3525759
- DOI: 10.1158/0008-5472.CAN-12-1656
An integrated genome-wide approach to discover tumor-specific antigens as potential immunologic and clinical targets in cancer
Abstract
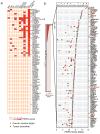
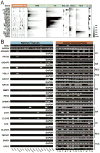
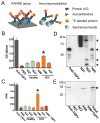
Tumor-specific antigens (TSA) are central elements in the immune control of cancers. To systematically explore the TSA genome, we developed a computational technology called heterogeneous expression profile analysis (HEPA), which can identify genes relatively uniquely expressed in cancer cells in contrast to normal somatic tissues. Rating human genes by their HEPA score enriched for clinically useful TSA genes, nominating candidate targets whose tumor-specific expression was verified by reverse transcription PCR (RT-PCR). Coupled with HEPA, we designed a novel assay termed protein A/G-based reverse serological evaluation (PARSE) for quick detection of serum autoantibodies against an array of putative TSA genes. Remarkably, highly tumor-specific autoantibody responses against seven candidate targets were detected in 4% to 11% of patients, resulting in distinctive autoantibody signatures in lung and stomach cancers. Interrogation of a larger cohort of 149 patients and 123 healthy individuals validated the predictive value of the autoantibody signature for lung cancer. Together, our results establish an integrated technology to uncover a cancer-specific antigen genome offering a reservoir of novel immunologic and clinical targets.
Conflict of interest statement
Figures


Similar articles
-
NY-ESO-1 Based Immunotherapy of Cancer: Current Perspectives.Front Immunol. 2018 May 1;9:947. doi: 10.3389/fimmu.2018.00947. eCollection 2018. Front Immunol. 2018. PMID: 29770138 Free PMC article. Review.
-
Tumor antigen discovery through translation of the cancer genome.Immunol Res. 2014 May;58(2-3):292-9. doi: 10.1007/s12026-014-8505-4. Immunol Res. 2014. PMID: 24718952 Review.
-
Genome-scale search of tumor-specific antigens by collective analysis of mutations, expressions and T-cell recognition.Mol Immunol. 2009 May;46(8-9):1824-9. doi: 10.1016/j.molimm.2009.01.019. Epub 2009 Feb 24. Mol Immunol. 2009. PMID: 19243822
-
Cancer immunotherapy using novel tumor-associated antigenic peptides identified by genome-wide cDNA microarray analyses.Cancer Sci. 2015 May;106(5):505-11. doi: 10.1111/cas.12650. Epub 2015 Apr 1. Cancer Sci. 2015. PMID: 25726868 Free PMC article. Review.
-
Predictive gene signature in MAGE-A3 antigen-specific cancer immunotherapy.J Clin Oncol. 2013 Jul 1;31(19):2388-95. doi: 10.1200/JCO.2012.44.3762. Epub 2013 May 28. J Clin Oncol. 2013. PMID: 23715562 Clinical Trial.
Cited by
-
Systematic identification of cancer-specific MHC-binding peptides with RAVEN.Oncoimmunology. 2018 Jul 23;7(9):e1481558. doi: 10.1080/2162402X.2018.1481558. eCollection 2018. Oncoimmunology. 2018. PMID: 30228952 Free PMC article.
-
CRISPR/Cas9-mediated target validation of the splicing inhibitor Pladienolide B.Biochim Open. 2016 Feb 24;3:72-75. doi: 10.1016/j.biopen.2016.02.001. eCollection 2016 Dec. Biochim Open. 2016. PMID: 29450134 Free PMC article.
-
Proteogenomic Characterization Reveals Therapeutic Vulnerabilities in Lung Adenocarcinoma.Cell. 2020 Jul 9;182(1):200-225.e35. doi: 10.1016/j.cell.2020.06.013. Cell. 2020. PMID: 32649874 Free PMC article.
-
Combination of OipA, BabA, and SabA as candidate biomarkers for predicting Helicobacter pylori-related gastric cancer.Sci Rep. 2016 Nov 7;6:36442. doi: 10.1038/srep36442. Sci Rep. 2016. PMID: 27819260 Free PMC article.
-
Role of Regenerating Islet-Derived Protein 3A in Gastrointestinal Cancer.Front Oncol. 2019 Dec 17;9:1449. doi: 10.3389/fonc.2019.01449. eCollection 2019. Front Oncol. 2019. PMID: 31921694 Free PMC article. Review.
References
-
- Gebo KA, Chander G, Jenckes MW, Ghanem KG, Herlong HF, Torbenson MS, et al. Screening tests for hepatocellular carcinoma in patients with chronic hepatitis C: a systematic review. Hepatology. 2002;36:S84–92. - PubMed
-
- Wang X, Yu J, Sreekumar A, Varambally S, Shen R, Giacherio D, et al. Autoantibody signatures in prostate cancer. N Engl J Med. 2005;353:1224–35. - PubMed
-
- Qiu J, Hanash S. Autoantibody profiling for cancer detection. Clin Lab Med. 2009;29:31–46. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

