Impact of strain typing methods on assessment of relationship between paired nares and wound isolates of methicillin-resistant Staphylococcus aureus
- PMID: 23135945
- PMCID: PMC3536221
- DOI: 10.1128/JCM.02423-12
Impact of strain typing methods on assessment of relationship between paired nares and wound isolates of methicillin-resistant Staphylococcus aureus
Abstract
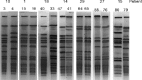
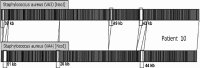
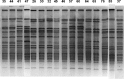
The anterior nares are the site of choice for the Veterans Administration methicillin-resistant Staphylococcus aureus (MRSA) surveillance program; however, a correlation between nares colonization and concomitant wound infections has not been well established. The purpose of this study was 3-fold: to determine the relatedness of MRSA isolates from 40 paired wound and nares specimens by four different strain typing methods, to determine concordance of typing methods, and to establish a baseline of MRSA types at this medical center. Isolates were typed by repetitive PCR (rep-PCR) (DiversiLab System; DL) and SpectraCell Raman analysis (SCRA) (commercially available methods that can be performed within a clinical lab), pulsed-field gel electrophoresis (PFGE), and an antibiotic susceptibility profile (AB). Whole-genome optical mapping (WGM) (OpGen, Inc.) was performed on selected isolates. All methods agreed that 26 pairs were indistinguishable and four pairs were different. Discrepant results were as follows: 4 where only SCRA was discordant, 3 where only AB was discordant, 2 where both DL and AB were discordant, and 1 where both DL and SCRA were discordant. All WGM agreed with PFGE. After discrepancy resolution, 80% of the pairs were indistinguishable and 20% were different. A total of 56% of nares results were nonpredictive if negative nares and positive wound cultures are included. Methods agreed 85 to 93% of the time; however, congruence of isolates to a clade was lower. Baseline analysis of types showed that 15 pairs were unique to single patients (30 strains, 38%; 47% of the matching pairs). Twenty-five strains (30%) represented a single clade identical by PFGE, SCRA, and DL, decreasing specificity. Typing method and institutional type frequency are important in assessing MRSA strain relatedness.
Figures
Similar articles
-
Comparison of typing results obtained for methicillin-resistant Staphylococcus aureus isolates with the DiversiLab system and pulsed-field gel electrophoresis.J Clin Microbiol. 2009 Aug;47(8):2452-7. doi: 10.1128/JCM.00476-09. Epub 2009 Jun 24. J Clin Microbiol. 2009. PMID: 19553588 Free PMC article.
-
Comparison of the DiversiLab repetitive element PCR system with spa typing and pulsed-field gel electrophoresis for clonal characterization of methicillin-resistant Staphylococcus aureus.J Clin Microbiol. 2011 Apr;49(4):1549-55. doi: 10.1128/JCM.02254-10. Epub 2011 Feb 9. J Clin Microbiol. 2011. PMID: 21307215 Free PMC article.
-
Evaluation of repetitive element polymerase chain reaction for surveillance of methicillin-resistant Staphylococcus aureus at a large academic medical center and community hospitals.Diagn Microbiol Infect Dis. 2015 Jan;81(1):13-7. doi: 10.1016/j.diagmicrobio.2014.05.005. Epub 2014 May 20. Diagn Microbiol Infect Dis. 2015. PMID: 25439582
-
Good performance of the SpectraCellRA system for typing of methicillin-resistant Staphylococcus aureus isolates.J Clin Microbiol. 2013 May;51(5):1434-8. doi: 10.1128/JCM.02101-12. Epub 2013 Feb 20. J Clin Microbiol. 2013. PMID: 23426926 Free PMC article.
-
Whole-genome mapping: a new paradigm in strain-typing technology.J Clin Microbiol. 2013 Apr;51(4):1066-70. doi: 10.1128/JCM.00093-13. Epub 2013 Jan 30. J Clin Microbiol. 2013. PMID: 23363821 Free PMC article. Review.
Cited by
-
Chrysanthemum zawadskii var. latilobum Flower Essential Oil Reduces MRSA Pathogenicity by Inhibiting Virulence Gene Expression.Molecules. 2025 Jan 25;30(3):553. doi: 10.3390/molecules30030553. Molecules. 2025. PMID: 39942657 Free PMC article.
-
In Vitro Activity of Retapamulin and Antimicrobial Susceptibility Patterns in a Longitudinal Collection of Methicillin-Resistant Staphylococcus aureus Isolates from a Veterans Affairs Medical Center.Antimicrob Agents Chemother. 2015 Dec 14;60(3):1298-303. doi: 10.1128/AAC.01568-15. Antimicrob Agents Chemother. 2015. PMID: 26666950 Free PMC article.
-
Investigation of a Candida guilliermondii Pseudo-outbreak Reveals a Novel Source of Laboratory Contamination.J Clin Microbiol. 2017 Apr;55(4):1080-1089. doi: 10.1128/JCM.02336-16. Epub 2017 Jan 18. J Clin Microbiol. 2017. PMID: 28100597 Free PMC article.
-
Novel MRSA-targeting phage MetB16: Genomic features, structural insights, and therapeutic applications.Turk J Biol. 2025 Feb 14;49(3):292-308. doi: 10.55730/1300-0152.2746. eCollection 2025. Turk J Biol. 2025. PMID: 40678418 Free PMC article.
-
Characterization of the resistome and predominant genetic lineages of Gram-positive bacteria causing keratitis.Antimicrob Agents Chemother. 2024 Mar 6;68(3):e0124723. doi: 10.1128/aac.01247-23. Epub 2024 Jan 30. Antimicrob Agents Chemother. 2024. PMID: 38289077 Free PMC article.
References
-
- Caffrey AR, Laplante KL. 2012. Changing epidemiology of methicillin-resistant Staphylococcus aureus in the Veterans Affairs Healthcare System, 2002-2009. Infection 40:291–297 - PubMed
-
- Jain R, Kralovic SM, Evans ME, Ambrose M, Simbartl LA, Obrosky DS, Render ML, Freyberg RW, Jernigan JA, Muder RR, Miller LJ, Roselle GA. 2011. Veterans Affairs initiative to prevent methicillin-resistant Staphylococcus aureus infections. N. Engl. J. Med. 364:1419–1430 - PubMed
-
- von Eiff C, Becker K, Machka K, Stammer H, Peters G. 2001. Nasal carriage as a source of Staphylococcus aureus bacteremia. N. Engl. J. Med. 344:11–16 - PubMed
-
- van Belkum A. 2007. Tracing isolates of bacterial species by multilocus variable number of tandem repeat analysis (MLVA). FEMS Immunol. Med. Microbiol. 49:22–27 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

