The role of electrostatic interactions on klentaq1 insight for domain separation
- PMID: 23136465
- PMCID: PMC3491847
- DOI: 10.4137/BBI.S9390
The role of electrostatic interactions on klentaq1 insight for domain separation
Abstract
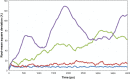
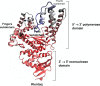
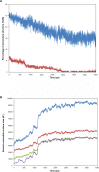
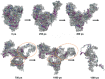
We investigated the relationship between the thermostability of Klentaq1 and factors stabilizing interdomain interactions. When thermal adaptation of Klentaq1 was analyzed at the atomic level, the protein was stable at 300 and 350 K. It gradually unfolded at 373 K and almost spontaneously unfolded at 400 K. Domain separation was induced by disrupting electrostatic interactions in two salt bridges formed by Lys354-Glu445 and Asp371-Arg435 on the interface domain. The role of these interactions in protein stability was evaluated by comparing free energy solvation (ΔΔG(solv)) between wild type and mutants. Substitution of Asp371 by Glu or Asn, and also Glu445 by Asn resulted in a positive value of ΔΔG(solv), suggesting that mutations destabilized the protein structure. Nevertheless, substitution of Glu445 by Asp gave a negative value to ΔΔG(solv) reflecting increasing protein stability. Our results demonstrate that interactions at the interface domains of Klentaq1 are essential factors correlated with the Klentaq1 thermostability.
Keywords: Klentaq1; domain separation; electrostatic interaction; in silico mutation.
Figures
References
-
- Rothwell PJ, Waksman G. Structure and mechanism of DNA polymerases. Adv Protein Chem. 2005;71:401–40. - PubMed
-
- Kornberg A, Baker TA. DNA Replication. 2nd ed. New Jersey: University Science Books; 2005.
LinkOut - more resources
Full Text Sources

