Molecular characterization of two high-palmitic-acid mutant loci induced by X-ray irradiation in soybean
- PMID: 23136502
- PMCID: PMC3406799
- DOI: 10.1270/jsbbs.61.631
Molecular characterization of two high-palmitic-acid mutant loci induced by X-ray irradiation in soybean
Abstract
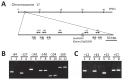
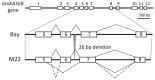
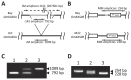
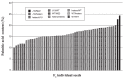
Palmitic acid is the most abundant (approx. 11% of total fatty acids) saturated fatty acid in conventional soybean seed oil. Increasing the saturated acid content of soybean oil improves its oxidative stability and plasticity. We have developed three soybean mutants with high palmitic acid content by X-ray irradiation. In this study, we successfully identified the mutated sites of two of these high-palmitic-acid mutants, J10 and M22. PCR-based mutant analysis revealed that J10 has a 206,203-bp-long deletion that includes the GmKASIIA gene and 16 other predicted genes, and M22 has a 26-bp-long deletion in the sixth intron of GmKASIIB. The small deletion in M22 causes mis-splicing of GmKASIIB transcripts, which should result in nonfunctional products. In addition, we designed co-dominant marker sets for these mutant alleles and confirmed the association of genotypes and palmitic acid contents in F(2) seeds of J10 X M22. This information will be useful in breeding programs to develop novel soybean cultivars with improved palmitic acid content. However, in the third mutant, KK7, we found no polymorphism in either GmKASIIA or GmKASIIB, which suggests that several unknown genes in addition to GmKASIIA and GmKASIIB may be involved in elevating the palmitic acid content of soybean seed oil.
Keywords: Glycine max; GmKASIIA; GmKASIIB; X-ray irradiation; palmitic acid.
Figures

References
-
- Aghoram K, Wilson RF, Burton JW, Dewey RE. A mutation in a 3-keto-acyl-ACP synthase gene is associated with elevated palmitic acid levels in soybean seeds. Crop Sci. 2006;46:2453–2459.
-
- Anai T, Yamada T, Kinoshita T, Rahman SM, Takagi Y. Identification of corresponding genes for three low-α-linolenic acid mutants and elucidation of their contribution to fatty acid biosynthesis in soybean seed. Plant Sci. 2005;168:1615–1623.
-
- Anai T, Yamada T, Hideshima R, Kinoshita T, Rahman SM, Takagi Y. Two high-oleic-acid soybean mutants, M23 and KK21, have disrupted microsomal omega-6 fatty acid desaturase, encoded by GmFAD2-1a. Breed Sci. 2008;58:447–452.
-
- Carlsson AS, LaBrie ST, Kinney AJ, von Wettstein-Knowles P, Browse J. A KAS2 cDNA complements the phenotypes of the Arabidopsis fab1 mutant that differs in a single residue bordering the substrate binding pocket. Plant J. 2002;29:761–770. - PubMed
-
- Erickson EA, Wilcox JR, Cavins JF. Inheritance of altered palmitic acid percentages in two soybean mutants. J Hered. 1988;79:465–468.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
