A Dual Reporter Splicing Assay Using HaloTag-containing Proteins
- PMID: 23136623
- PMCID: PMC3486960
- DOI: 10.2174/1875397301206010027
A Dual Reporter Splicing Assay Using HaloTag-containing Proteins
Abstract
To evaluate the effects of genetic variations on mRNA splicing, we developed a minigene-based splicing assay using reporter genes encoding luciferase and the multifunctional HaloTag protein. In addition to conventional RT-PCR analysis, splicing events can be monitored in this system using two parameters: luciferase activity and signals derived from HaloTag-containing proteins bound to a fluorescent ligand following SDS-PAGE. The luciferase activity reflects the accumulated amounts of successfully spliced HaloTag-luciferase fusion products, whereas the amounts and sizes of HaloTag-containing proteins provide quantitative insights into precursor, correctly spliced, and aberrantly spliced mRNA species. Preliminary experiments confirmed that the dual reporter minigene assay can provide estimates of overall splicing efficiency based on the levels of protein products. We then used the minigene assay to analyze a case of chronic granulomatous disease that was caused by a G>C mutation at position +5 in the 5'-splice donor site of intron 5 of the CYBB gene. We found that the G>C mutation affected CYBB mRNA splicing by changing a delicate balance of splicing efficiencies of introns 4, 5, and 6.
Keywords: CYBB gene; Chronic granulomatous disease; HaloTag fusion protein; genetic mutation.; luciferase reporter assay; splicing.
Figures
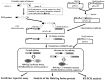


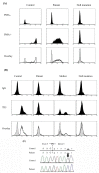




Similar articles
-
Activation of cryptic splice sites in three patients with chronic granulomatous disease.Mol Genet Genomic Med. 2019 Sep;7(9):e854. doi: 10.1002/mgg3.854. Epub 2019 Jul 30. Mol Genet Genomic Med. 2019. PMID: 31364312 Free PMC article.
-
Interferon-gamma improves splicing efficiency of CYBB gene transcripts in an interferon-responsive variant of chronic granulomatous disease due to a splice site consensus region mutation.Blood. 2000 Jun 1;95(11):3548-54. Blood. 2000. PMID: 10828042
-
In vitro splicing analysis showed that availability of a cryptic splice site is not a determinant for alternative splicing patterns caused by +1G-->A mutations in introns of the dystrophin gene.J Med Genet. 2009 Aug;46(8):542-7. doi: 10.1136/jmg.2008.061259. Epub 2008 Nov 10. J Med Genet. 2009. PMID: 19001018
-
Principles and Practical Considerations for the Analysis of Disease-Associated Alternative Splicing Events Using the Gateway Cloning-Based Minigene Vectors pDESTsplice and pSpliceExpress.Int J Mol Sci. 2021 May 13;22(10):5154. doi: 10.3390/ijms22105154. Int J Mol Sci. 2021. PMID: 34068052 Free PMC article. Review.
-
Cancer-Associated Perturbations in Alternative Pre-messenger RNA Splicing.Cancer Treat Res. 2013;158:41-94. doi: 10.1007/978-3-642-31659-3_3. Cancer Treat Res. 2013. PMID: 24222354 Review.
Cited by
-
HaloTag technology: a versatile platform for biomedical applications.Bioconjug Chem. 2015 Jun 17;26(6):975-86. doi: 10.1021/acs.bioconjchem.5b00191. Epub 2015 May 22. Bioconjug Chem. 2015. PMID: 25974629 Free PMC article. Review.
-
Rational Design of an Activatable Reporter for Quantitative Imaging of RNA Aberrant Splicing In Vivo.Mol Ther Methods Clin Dev. 2020 Apr 18;17:904-911. doi: 10.1016/j.omtm.2020.04.007. eCollection 2020 Jun 12. Mol Ther Methods Clin Dev. 2020. PMID: 32405512 Free PMC article.
-
HaloTag® Platform: From Proteomics to Cellular Analysis and Animal Imaging.Curr Chem Genomics. 2012;6:6-7. doi: 10.2174/1875397301206010006. Epub 2012 Sep 20. Curr Chem Genomics. 2012. PMID: 23115609 Free PMC article. No abstract available.
References
-
- Cooper TA. Use of minigene systems to dissect alternative splicing elements. Methods. 2005;37:331–40. - PubMed
-
- Singh G, Cooper TA. Minigene reporter for identification and analysis of cis elements and trans factors affecting pre-mRNA splicing. Biotechniques. 2006;41:177–81. - PubMed
-
- Andreassi C, Jarecki J, Zhou J, et al. Aclarubicin treatment restores SMN levels to cells derived from type I spinal muscular atrophy patients. Hum Mol Genet. 2001;10:2841–9. - PubMed
-
- Los GV, Wood K. The HaloTag: a novel technology for cell imaging and protein analysis. Methods Mol Biol. 2007;356:195–208. - PubMed
-
- Ho SN, Hunt HD, Horton RM, et al. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–9. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
