Platelet-derived growth factors and their receptors: structural and functional perspectives
- PMID: 23137658
- PMCID: PMC3612563
- DOI: 10.1016/j.bbapap.2012.10.015
Platelet-derived growth factors and their receptors: structural and functional perspectives
Abstract
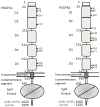
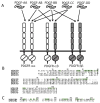
The four types of platelet-derived growth factors (PDGFs) and the two types of PDGF receptors (PDGFRs, which belong to class III receptor tyrosine kinases) have important functions in the development of connective tissue cells. Recent structural studies have revealed novel mechanisms of PDGFs in propeptide loading and receptor recognition/activation. The detailed structural understanding of PDGF-PDGFR signaling has provided a template that can aid therapeutic intervention to counteract the aberrant signaling of this normally silent pathway, especially in proliferative diseases such as cancer. This review summarizes the advances in the PDGF system with a focus on relating the structural and functional understandings, and discusses the basic aspects of PDGFs and PDGFRs, the mechanisms of activation, and the insights into the therapeutic antagonism of PDGFRs. This article is part of a Special Issue entitled: Emerging recognition and activation mechanisms of receptor tyrosine kinases.
Keywords: Growth factor; Propeptide recognition; Receptor activation; Receptor tyrosine kinase; Signal transduction.
Copyright © 2012 Elsevier B.V. All rights reserved.
Figures



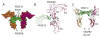

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

