Transfer RNA modifications: nature's combinatorial chemistry playground
- PMID: 23139145
- PMCID: PMC3680101
- DOI: 10.1002/wrna.1144
Transfer RNA modifications: nature's combinatorial chemistry playground
Abstract
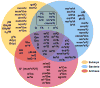
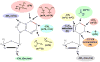
Following synthesis, tRNAs are peppered by numerous chemical modifications which may differentially affect a tRNA's structure and function. Although modifications affecting the business ends of a tRNA are predictably important for cell viability, a majority of modifications play more subtle structural roles that can affect tRNA stability and folding. The current trend is that modifications act in concert and it is in the context of the specific sequence of a given tRNA that they impart their differing effects. Recent developments in the modification field have highlighted the diversity of modifications in tRNA. From these, the combinatorial nature of modifications in explaining previously described phenotypes derived from their absence has emerged as a growing theme.
Copyright © 2012 John Wiley & Sons, Ltd.
Figures
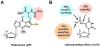

References
-
- Grosjean H. DNA and RNA modification enzymes: structure, mechanism, function and evolution. Austin, Tex: Landes Bioscience; 2009.
-
- Phillips G, de Crecy-Lagard V. Biosynthesis and function of tRNA modifications in Archaea. Curr Opin Microbiol. 2011;14:335–341. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

