Multiple sodium channel variants in the mosquito Culex quinquefasciatus
- PMID: 23139629
- PMCID: PMC3492789
- DOI: 10.7150/ijbs.4966
Multiple sodium channel variants in the mosquito Culex quinquefasciatus
Abstract
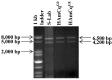
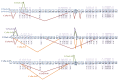
Voltage-gated sodium channels are the target sites of both DDT and pyrethroid insecticides. The importance of alternative splicing as a key mechanism governing the structural and functional diversity of sodium channels and the resulting development of insecticide and acaricide resistance is widely recognized, as shown by the extensive research on characterizing alternative splicing and variants of sodium channels in medically and agriculturally important insect species. Here we present the first comparative study of multiple variants of the sodium channel transcripts in the mosquito Culex quinquefasciatus. The variants were classified into two categories, CxNa-L and CxNa-S based on their distinguishing sequence sizes of ~6.5 kb and ~4.0 kb, respectively, and generated via major extensive alternative splicing with minor small deletions/ insertions in susceptible S-Lab, low resistant HAmCq(G0), and highly resistant HAmCq(G8)Culex strains. Four alternative Cx-Na-L splice variants were identified, including three full length variants with three optional exons (2, 5, and 21i) and one with in-frame-stop codons. Large, multi-exon-alternative splices were identified in the CxNa-S category. All CxNa-S splicing variants in the S-Lab and HAmCq(G0) strains contained in-frame stop codons, suggesting that any resulting proteins would be truncated. The ~1000 to ~3000-fold lower expression of these splice variants with stop codons compared with the CxNa-L splicing variants may support the lower importance of these variants in S-Lab and HAmCq(G0). Interestingly, two alternative splicing variants of CxNa-S in HAmCq(G8) included entire ORFs but lacked exons 5 to18 and these two variants had much higher expression levels in HAmCq(G8) than in S-Lab and HAmCq(G0). These results provide a functional basis for further characterizing how alternative splicing of a voltage-gated sodium channel contributes to diversity in neuronal signaling in mosquitoes in response to pyrethroids, and possibly indicates the role of these variants in the development of pyrethroid resistance.
Keywords: Culex quinquefasciatus.; Sodium channel; alternative splicing; insecticide resistance; transcript variants.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures











Similar articles
-
Permethrin resistance variation and susceptible reference line isolation in a field population of the mosquito, Culex quinquefasciatus (Diptera: Culicidae).Insect Sci. 2014 Oct;21(5):659-66. doi: 10.1111/1744-7917.12071. Epub 2013 Dec 4. Insect Sci. 2014. PMID: 24357606
-
Genome analysis of cytochrome P450s and their expression profiles in insecticide resistant mosquitoes, Culex quinquefasciatus.PLoS One. 2011;6(12):e29418. doi: 10.1371/journal.pone.0029418. Epub 2011 Dec 29. PLoS One. 2011. PMID: 22242119 Free PMC article.
-
Resistance in the mosquito, Culex quinquefasciatus, and possible mechanisms for resistance.Pest Manag Sci. 2005 Nov;61(11):1096-102. doi: 10.1002/ps.1090. Pest Manag Sci. 2005. PMID: 16032654
-
Life and Death at the Voltage-Sensitive Sodium Channel: Evolution in Response to Insecticide Use.Annu Rev Entomol. 2019 Jan 7;64:243-257. doi: 10.1146/annurev-ento-011118-112420. Annu Rev Entomol. 2019. PMID: 30629893 Review.
-
Pyrethroid resistance in Culex pipiens mosquitoes.Pestic Biochem Physiol. 2015 May;120:68-76. doi: 10.1016/j.pestbp.2014.12.018. Epub 2014 Dec 19. Pestic Biochem Physiol. 2015. PMID: 25987223 Review.
Cited by
-
Sodium channel point mutations associated with pyrethroid resistance in Chinese strains of Culex pipiens quinquefasciatus (Diptera: Culicidae).Parasit Vectors. 2014 Aug 15;7:369. doi: 10.1186/1756-3305-7-369. Parasit Vectors. 2014. PMID: 25128988 Free PMC article.
-
Cloning of multiple ERα mRNA variants in killifish (Fundulus heteroclitus), and differential expression by tissue type, stage of reproduction, and estrogen exposure in fish from polluted and unpolluted environments.Aquat Toxicol. 2015 Feb;159:184-97. doi: 10.1016/j.aquatox.2014.12.012. Epub 2014 Dec 18. Aquat Toxicol. 2015. PMID: 25550165 Free PMC article.
-
Splice form variant and amino acid changes in MDR49 confers DDT resistance in transgenic Drosophila.Sci Rep. 2016 Mar 22;6:23355. doi: 10.1038/srep23355. Sci Rep. 2016. PMID: 27003579 Free PMC article.
-
Detection of the Nav channel kdr-like mutation and modeling of factors affecting survivorship of Culex quinquefasciatus mosquitoes from six areas of Harris County (Houston), Texas, after permethrin field-cage tests.PLoS Negl Trop Dis. 2020 Nov 19;14(11):e0008860. doi: 10.1371/journal.pntd.0008860. eCollection 2020 Nov. PLoS Negl Trop Dis. 2020. PMID: 33211688 Free PMC article.
-
Using targeted next-generation sequencing to characterize genetic differences associated with insecticide resistance in Culex quinquefasciatus populations from the southern U.S.PLoS One. 2019 Jul 3;14(7):e0218397. doi: 10.1371/journal.pone.0218397. eCollection 2019. PLoS One. 2019. PMID: 31269040 Free PMC article.
References
-
- Bloomquist JR. Ion channels as targets for insecticides. Annu Rev Entomol. 1996;41:163–190. - PubMed
-
- Narahashi T. Neuronal ion channels as the target sites of insecticides. Pharmacol Toxico. 1996;7:1–14. - PubMed
-
- Soderlund DM. Sodium channels. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive molecular insect science. New York: Elsevier Pergamon; 2005. pp. 1–24.
-
- Soderlund DM, Knipple DC. The molecular biology of knockdown resistance to pyrethroid insecticides. Insect Biochem Mol Biol. 2003;33:563–77. - PubMed
-
- Catterall WA. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron. 2000;26:13–25. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

