Microfluidic systems for single DNA dynamics
- PMID: 23139700
- PMCID: PMC3489478
- DOI: 10.1039/C2SM26036K
Microfluidic systems for single DNA dynamics
Abstract
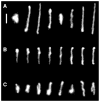
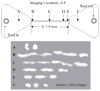
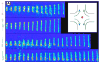
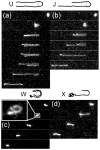
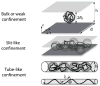
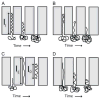
Recent advances in microfluidics have enabled the molecular-level study of polymer dynamics using single DNA chains. Single polymer studies based on fluorescence microscopy allow for the direct observation of non-equilibrium polymer conformations and dynamical phenomena such as diffusion, relaxation, and molecular stretching pathways in flow. Microfluidic devices have enabled the precise control of model flow fields to study the non-equilibrium dynamics of soft materials, with device geometries including curved channels, cross-slots, and microfabricated obstacles and structures. This review explores recent microfluidic systems that have advanced the study of single polymer dynamics, while identifying new directions in the field that will further elucidate the relationship between polymer microstructure and bulk rheological properties.
Figures
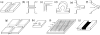

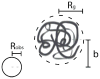

References
Grants and funding
LinkOut - more resources
Full Text Sources

