G-cimp status prediction of glioblastoma samples using mRNA expression data
- PMID: 23139755
- PMCID: PMC3490960
- DOI: 10.1371/journal.pone.0047839
G-cimp status prediction of glioblastoma samples using mRNA expression data
Abstract
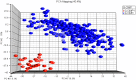
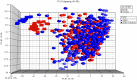
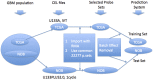
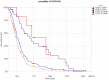
Glioblastoma Multiforme (GBM) is a tumor with high mortality and no known cure. The dramatic molecular and clinical heterogeneity seen in this tumor has led to attempts to define genetically similar subgroups of GBM with the hope of developing tumor specific therapies targeted to the unique biology within each of these subgroups. Recently, a subset of relatively favorable prognosis GBMs has been identified. These glioma CpG island methylator phenotype, or G-CIMP tumors, have distinct genomic copy number aberrations, DNA methylation patterns, and (mRNA) expression profiles compared to other GBMs. While the standard method for identifying G-CIMP tumors is based on genome-wide DNA methylation data, such data is often not available compared to the more widely available gene expression data. In this study, we have developed and evaluated a method to predict the G-CIMP status of GBM samples based solely on gene expression data.
Conflict of interest statement
Figures

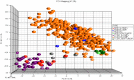

References
-
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, et al. (2006) Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer cell 9: 157–173. - PubMed
-
- Huse JT, Phillips HS, Brennan CW (2011) Molecular subclassification of diffuse gliomas: seeing order in the chaos. Glia. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

