Structure-function of the G protein-coupled receptor superfamily
- PMID: 23140243
- PMCID: PMC3540149
- DOI: 10.1146/annurev-pharmtox-032112-135923
Structure-function of the G protein-coupled receptor superfamily
Abstract
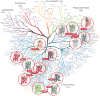
During the past few years, crystallography of G protein-coupled receptors (GPCRs) has experienced exponential growth, resulting in the determination of the structures of 16 distinct receptors-9 of them in 2012 alone. Including closely related subtype homology models, this coverage amounts to approximately 12% of the human GPCR superfamily. The adrenergic, rhodopsin, and adenosine receptor systems are also described by agonist-bound active-state structures, including a structure of the receptor-G protein complex for the β(2)-adrenergic receptor. Biochemical and biophysical techniques, such as nuclear magnetic resonance and hydrogen-deuterium exchange coupled with mass spectrometry, are providing complementary insights into ligand-dependent dynamic equilibrium between different functional states. Additional details revealed by high-resolution structures illustrate the receptors as allosteric machines that are controlled not only by ligands but also by ions, lipids, cholesterol, and water. This wealth of data is helping redefine our knowledge of how GPCRs recognize such a diverse array of ligands and how they transmit signals 30 angstroms across the cell membrane; it also is shedding light on a structural basis of GPCR allosteric modulation and biased signaling.
Figures




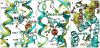

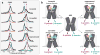
References
-
- Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–57. - PubMed
-
- Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–72. - PubMed
-
- Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–6. - PubMed
-
- Tyndall JD, Sandilya R. GPCR agonists and antagonists in the clinic. Med Chem. 2005;1:405–21. - PubMed
-
- Lappano R, Maggiolini M. G protein-coupled receptors: novel targets for drug discovery in cancer. Nat Rev Drug Discov. 2011;10:47–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

