Nucleosomal elements that control the topography of the barrier to transcription
- PMID: 23141536
- PMCID: PMC3508686
- DOI: 10.1016/j.cell.2012.10.009
Nucleosomal elements that control the topography of the barrier to transcription
Abstract
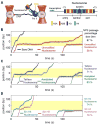
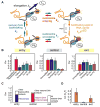
The nucleosome represents a mechanical barrier to transcription that operates as a general regulator of gene expression. We investigate how each nucleosomal component-the histone tails, the specific histone-DNA contacts, and the DNA sequence-contributes to the strength of the barrier. Removal of the tails favors progression of RNA polymerase II into the entry region of the nucleosome by locally increasing the wrapping-unwrapping rates of the DNA around histones. In contrast, point mutations that affect histone-DNA contacts at the dyad abolish the barrier to transcription in the central region by decreasing the local wrapping rate. Moreover, we show that the nucleosome amplifies sequence-dependent transcriptional pausing, an effect mediated through the structure of the nascent RNA. Each of these nucleosomal elements controls transcription elongation by affecting distinctly the density and duration of polymerase pauses, thus providing multiple and alternative mechanisms for control of gene expression by chromatin remodeling and transcription factors.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures



References
-
- Anderson J, Lowary P, Widom J. Effects of histone acetylation on the equilibrium accessibility of nucleosomal DNA target sites1. J Mol Biol. 2001;307:977–985. - PubMed
-
- Bai L, Shundrovsky A, Wang MD. Sequence-dependent kinetic model for transcription elongation by RNA polymerase. J Mol Biol. 2004;344:335–349. - PubMed
-
- Bondarenko VA, Steele LM, Újvári A, Gaykalova DA, Kulaeva OI, Polikanov YS, Luse DS, Studitsky VM. Nucleosomes Can Form a Polar Barrier to Transcript Elongation by RNA Polymerase II. Mol Cell. 2006;24:469–479. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

