Modulation of hepatitis C virus RNA abundance and virus release by dispersion of processing bodies and enrichment of stress granules
- PMID: 23141719
- PMCID: PMC3534916
- DOI: 10.1016/j.virol.2012.10.027
Modulation of hepatitis C virus RNA abundance and virus release by dispersion of processing bodies and enrichment of stress granules
Abstract
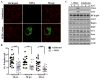
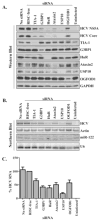
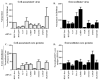
Components of cytoplasmic processing bodies (P-bodies) and stress granules can be subverted during viral infections to modulate viral gene expression. Because hepatitis C virus (HCV) RNA abundance is regulated by P-body components such as microRNA miR-122, Argonaute 2 and RNA helicase RCK/p54, we examined whether HCV infection modulates P-bodies and stress granules during viral infection. It was discovered that HCV infection decreased the number of P-bodies, but induced the formation of stress granules. Immunofluorescence studies revealed that a number of P-body and stress granule proteins co-localized with viral core protein at lipid droplets, the sites for viral RNA packaging. Depletion of selected P-body proteins decreased overall HCV RNA and virion abundance. Depletion of stress granule proteins also decreased overall HCV RNA abundance, but surprisingly enhanced the accumulation of infectious, extracellular virus. These data argue that HCV subverts P-body and stress granule components to aid in viral gene expression at particular sites in the cytoplasm.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures





References
-
- Adler J, Parmryd I. Quantifying colocalization by correlation: the Pearson correlation coefficient is superior to the Mander’s overlap coefficient. Cytometry A. 2010;77:733–742. - PubMed
-
- Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009a;10:430–436. - PubMed
-
- Anderson P, Kedersha N. Stress granules. Curr Biol. 2009b;19:R397–398. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

