The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response
- PMID: 23142775
- PMCID: PMC3501571
- DOI: 10.1038/ni.2460
The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response
Abstract
The induction of type I interferons by the bacterial secondary messengers cyclic di-GMP (c-di-GMP) or cyclic di-AMP (c-di-AMP) is dependent on a signaling axis that involves the adaptor STING, the kinase TBK1 and the transcription factor IRF3. Here we identified the heliase DDX41 as a pattern-recognition receptor (PRR) that sensed both c-di-GMP and c-di-AMP. DDX41 specifically and directly interacted with c-di-GMP. Knockdown of DDX41 via short hairpin RNA in mouse or human cells inhibited the induction of genes encoding molecules involved in the innate immune response and resulted in defective activation of STING, TBK1 and IRF3 in response to c-di-GMP or c-di-AMP. Our results suggest a mechanism whereby c-di-GMP and c-di-AMP are detected by DDX41, which forms a complex with STING to signal to TBK1-IRF3 and activate the interferon response.
Figures

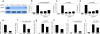
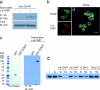


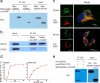
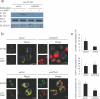
Comment in
-
Innate sensing of bacterial cyclic dinucleotides: more than just STING.Nat Immunol. 2012 Dec;13(12):1137-9. doi: 10.1038/ni.2469. Nat Immunol. 2012. PMID: 23160208 No abstract available.
Similar articles
-
Innate sensing of bacterial cyclic dinucleotides: more than just STING.Nat Immunol. 2012 Dec;13(12):1137-9. doi: 10.1038/ni.2469. Nat Immunol. 2012. PMID: 23160208 No abstract available.
-
The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides.Infect Immun. 2011 Feb;79(2):688-94. doi: 10.1128/IAI.00999-10. Epub 2010 Nov 22. Infect Immun. 2011. PMID: 21098106 Free PMC article.
-
c-di-GMP Induces COX-2 Expression in Macrophages in a STING-Independent Manner.ACS Chem Biol. 2021 Sep 17;16(9):1663-1670. doi: 10.1021/acschembio.1c00342. Epub 2021 Sep 3. ACS Chem Biol. 2021. PMID: 34478263
-
Cyclic di-nucleotide signaling enters the eukaryote domain.IUBMB Life. 2013 Nov;65(11):897-903. doi: 10.1002/iub.1212. Epub 2013 Oct 17. IUBMB Life. 2013. PMID: 24136904 Free PMC article. Review.
-
Cyclic di-GMP: second messenger extraordinaire.Nat Rev Microbiol. 2017 May;15(5):271-284. doi: 10.1038/nrmicro.2016.190. Epub 2017 Feb 6. Nat Rev Microbiol. 2017. PMID: 28163311 Review.
Cited by
-
Modulation of Clr Ligand Expression and NKR-P1 Receptor Function during Murine Cytomegalovirus Infection.J Innate Immun. 2015;7(6):584-600. doi: 10.1159/000382032. Epub 2015 May 29. J Innate Immun. 2015. PMID: 26044139 Free PMC article.
-
Bmp8a is an essential positive regulator of antiviral immunity in zebrafish.Commun Biol. 2021 Mar 9;4(1):318. doi: 10.1038/s42003-021-01811-0. Commun Biol. 2021. PMID: 33750893 Free PMC article.
-
Bacterial recognition pathways that lead to inflammasome activation.Immunol Rev. 2015 May;265(1):112-29. doi: 10.1111/imr.12289. Immunol Rev. 2015. PMID: 25879288 Free PMC article. Review.
-
Ddx41 inhibition of DNA damage signaling permits erythroid progenitor expansion in zebrafish.Haematologica. 2022 Mar 1;107(3):644-654. doi: 10.3324/haematol.2020.257246. Haematologica. 2022. PMID: 33763998 Free PMC article.
-
Intranasal vaccination with a plant-derived H5 HA vaccine protects mice and ferrets against highly pathogenic avian influenza virus challenge.Hum Vaccin Immunother. 2015;11(5):1235-43. doi: 10.4161/21645515.2014.988554. Hum Vaccin Immunother. 2015. PMID: 25714901 Free PMC article.
References
-
- McCoy CE, O'Neill LA. The role of toll-like receptors in macrophages. Front Biosci. 2008;13:62–70. - PubMed
-
- Pluddemann A, Mukhopadhyay S, Gordon S. Innate immunity to intracellular pathogens: macrophage receptors and responses to microbial entry. Immunol Rev. 2011;240:11–24. - PubMed
-
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–26. - PubMed
-
- Mills E, Pultz IS, Kulasekara HD, Miller SI. The bacterial second messenger c-di-GMP: mechanisms of signalling. Cell Microbiol. 2011;13:1122–9. - PubMed
-
- Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–73. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U19 AI067769/AI/NIAID NIH HHS/United States
- T32-AR058921/AR/NIAMS NIH HHS/United States
- R37 AI047868/AI/NIAID NIH HHS/United States
- R01 AI069120/AI/NIAID NIH HHS/United States
- AI056154/AI/NIAID NIH HHS/United States
- R01 AI022553/AI/NIAID NIH HHS/United States
- R01 AI056154/AI/NIAID NIH HHS/United States
- T32 AR058921/AR/NIAMS NIH HHS/United States
- R01 AI073539/AI/NIAID NIH HHS/United States
- R01 AI047868/AI/NIAID NIH HHS/United States
- AI067769/AI/NIAID NIH HHS/United States
- AR63020/AR/NIAMS NIH HHS/United States
- AI047868/AI/NIAID NIH HHS/United States
- P50 AR063020/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

