Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1
- PMID: 23142980
- PMCID: PMC3603146
- DOI: 10.1038/nsmb.2435
Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1
Abstract
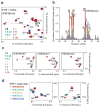
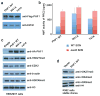
The PHD finger protein 1 (PHF1) is essential in epigenetic regulation and genome maintenance. Here we show that the Tudor domain of human PHF1 binds to histone H3 trimethylated at Lys36 (H3K36me3). We report a 1.9-Å resolution crystal structure of the Tudor domain in complex with H3K36me3 and describe the molecular mechanism of H3K36me3 recognition using NMR. Binding of PHF1 to H3K36me3 inhibits the ability of the Polycomb PRC2 complex to methylate Lys27 of histone H3 in vitro and in vivo. Laser microirradiation data show that PHF1 is transiently recruited to DNA double-strand breaks, and PHF1 mutants impaired in the H3K36me3 interaction exhibit reduced retention at double-strand break sites. Together, our findings suggest that PHF1 can mediate deposition of the repressive H3K27me3 mark and acts as a cofactor in early DNA-damage response.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




Comment in
-
H3K36me3 key to Polycomb-mediated gene silencing in lineage specification.Nat Struct Mol Biol. 2012 Dec;19(12):1214-5. doi: 10.1038/nsmb.2458. Nat Struct Mol Biol. 2012. PMID: 23211767 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

