Mechanism of promoter repression by Lac repressor-DNA loops
- PMID: 23143103
- PMCID: PMC3592455
- DOI: 10.1093/nar/gks1011
Mechanism of promoter repression by Lac repressor-DNA loops
Erratum in
- Nucleic Acids Res. 2013 Apr 1;41(6):3962. Lionberger, Troy A [added]
Abstract
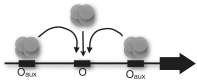
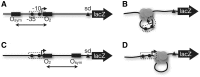
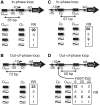
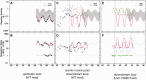
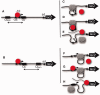
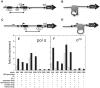
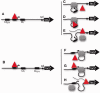
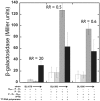
The Escherichia coli lactose (lac) operon encodes the first genetic switch to be discovered, and lac remains a paradigm for studying negative and positive control of gene expression. Negative control is believed to involve competition of RNA polymerase and Lac repressor for overlapping binding sites. Contributions to the local Lac repressor concentration come from free repressor and repressor delivered to the operator from remote auxiliary operators by DNA looping. Long-standing questions persist concerning the actual role of DNA looping in the mechanism of promoter repression. Here, we use experiments in living bacteria to resolve four of these questions. We show that the distance dependence of repression enhancement is comparable for upstream and downstream auxiliary operators, confirming the hypothesis that repressor concentration increase is the principal mechanism of repression loops. We find that as few as four turns of DNA can be constrained in a stable loop by Lac repressor. We show that RNA polymerase is not trapped at repressed promoters. Finally, we show that constraining a promoter in a tight DNA loop is sufficient for repression even when promoter and operator do not overlap.
Figures
Similar articles
-
Bacterial promoter repression by DNA looping without protein-protein binding competition.Nucleic Acids Res. 2014 May;42(9):5495-504. doi: 10.1093/nar/gku180. Epub 2014 Mar 5. Nucleic Acids Res. 2014. PMID: 24598256 Free PMC article.
-
Gene repression by minimal lac loops in vivo.Nucleic Acids Res. 2010 Dec;38(22):8072-82. doi: 10.1093/nar/gkq755. Nucleic Acids Res. 2010. PMID: 21149272 Free PMC article.
-
Engineered transcription activator-like effector dimer proteins confer DNA loop-dependent gene repression comparable to Lac repressor.Nucleic Acids Res. 2024 Sep 9;52(16):9996-10004. doi: 10.1093/nar/gkae656. Nucleic Acids Res. 2024. PMID: 39077947 Free PMC article.
-
The function of auxiliary operators.Mol Microbiol. 1998 Jul;29(1):13-8. doi: 10.1046/j.1365-2958.1998.00870.x. Mol Microbiol. 1998. PMID: 9701798 Review.
-
DNA looping in prokaryotes: experimental and theoretical approaches.J Bacteriol. 2013 Mar;195(6):1109-19. doi: 10.1128/JB.02038-12. Epub 2013 Jan 4. J Bacteriol. 2013. PMID: 23292776 Free PMC article. Review.
Cited by
-
Bacterial gene control by DNA looping using engineered dimeric transcription activator like effector (TALE) proteins.Nucleic Acids Res. 2018 Mar 16;46(5):2690-2696. doi: 10.1093/nar/gky047. Nucleic Acids Res. 2018. PMID: 29390154 Free PMC article.
-
Bacterial promoter repression by DNA looping without protein-protein binding competition.Nucleic Acids Res. 2014 May;42(9):5495-504. doi: 10.1093/nar/gku180. Epub 2014 Mar 5. Nucleic Acids Res. 2014. PMID: 24598256 Free PMC article.
-
Quantitative Determination of DNA Bridging Efficiency of Chromatin Proteins.Methods Mol Biol. 2024;2819:443-454. doi: 10.1007/978-1-0716-3930-6_20. Methods Mol Biol. 2024. PMID: 39028518
-
Transcription Attenuation in Synthetic Promoters in Nonoverlapping Tandem Formation.Biochemistry. 2024 Aug 20;63(16):2009-2022. doi: 10.1021/acs.biochem.4c00012. Epub 2024 Jul 12. Biochemistry. 2024. PMID: 38997112 Free PMC article.
-
Deep flanking sequence engineering for efficient promoter design using DeepSEED.Nat Commun. 2023 Oct 9;14(1):6309. doi: 10.1038/s41467-023-41899-y. Nat Commun. 2023. PMID: 37813854 Free PMC article.
References
-
- Müller-Hill B. The Lac Operon: A Short History of a Genetic Paradigm. Berlin, New York: Walter de Gruyter; 1996.
-
- Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961;3:318–356. - PubMed

