A multi-parametric flow cytometric assay to analyze DNA-protein interactions
- PMID: 23143268
- PMCID: PMC3554230
- DOI: 10.1093/nar/gks1034
A multi-parametric flow cytometric assay to analyze DNA-protein interactions
Abstract
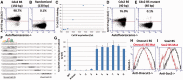
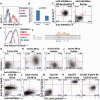
Interactions between DNA and transcription factors (TFs) guide cellular function and development, yet the complexities of gene regulation are still far from being understood. Such understanding is limited by a paucity of techniques with which to probe DNA-protein interactions. We have devised magnetic protein immobilization on enhancer DNA (MagPIE), a simple, rapid, multi-parametric assay using flow cytometric immunofluorescence to reveal interactions among TFs, chromatin structure and DNA. In MagPIE, synthesized DNA is bound to magnetic beads, which are then incubated with nuclear lysate, permitting sequence-specific binding by TFs, histones and methylation by native lysate factors that can be optionally inhibited with small molecules. Lysate protein-DNA binding is monitored by flow cytometric immunofluorescence, which allows for accurate comparative measurement of TF-DNA affinity. Combinatorial fluorescent staining allows simultaneous analysis of sequence-specific TF-DNA interaction and chromatin modification. MagPIE provides a simple and robust method to analyze complex epigenetic interactions in vitro.
Figures


References
-
- Koch F, Jourquin F, Ferrier P, Andrau JC. Genome-wide RNA polymerase II: not genes only! Trends Biochem. Sci. 2008;33:265–273. - PubMed
-
- Kee BL, Arias J, Montminy MR. Adaptor-mediated recruitment of RNA polymerase II to a signal-dependent activator. J. Biol. Chem. 1996;271:2373–2375. - PubMed
-
- Rundlett SE, Carmen AA, Suka N, Turner BM, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. - PubMed
-
- Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 1998;67:545–579. - PubMed
-
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

