Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes
- PMID: 23143334
- PMCID: PMC3631562
- DOI: 10.1038/nature11625
Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes
Abstract
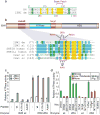
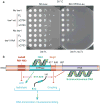
Chromatin-remodelling complexes (CRCs) mobilize nucleosomes to mediate the access of DNA-binding factors to their sites in vivo. These CRCs contain a catalytic subunit that bears an ATPase/DNA-translocase domain and flanking regions that bind nucleosomal epitopes. A central question is whether and how these flanking regions regulate ATP hydrolysis or the coupling of hydrolysis to DNA translocation, to affect nucleosome-sliding efficiency. ISWI-family CRCs contain the protein ISWI, which uses its ATPase/DNA-translocase domain to pump DNA around the histone octamer to enable sliding. ISWI is positively regulated by two 'activating' nucleosomal epitopes: the 'basic patch' on the histone H4 tail, and extranucleosomal (linker) DNA. Previous work defined the HAND-SANT-SLIDE (HSS) domain at the ISWI carboxy terminus that binds linker DNA, needed for ISWI activity. Here we define two new, conserved and separate regulatory regions on Drosophila ISWI, termed AutoN and NegC, which negatively regulate ATP hydrolysis (AutoN) or the coupling of ATP hydrolysis to productive DNA translocation (NegC). The two aforementioned nucleosomal epitopes promote remodelling indirectly by preventing the negative regulation of AutoN and NegC. Notably, mutation or removal of AutoN and NegC enables marked nucleosome sliding without the H4 basic patch or extranucleosomal DNA, or the HSS domain, conferring on ISWI the biochemical attributes normally associated with SWI/SNF-family ATPases. Thus, the ISWI ATPase catalytic core is an intrinsically active DNA translocase that conducts nucleosome sliding, onto which selective 'inhibition-of-inhibition' modules are placed, to help ensure that remodelling occurs only in the presence of proper nucleosomal epitopes. This supports a general concept for the specialization of chromatin-remodelling ATPases, in which specific regulatory modules adapt an ancient active DNA translocase to conduct particular tasks only on the appropriate chromatin landscape.
Figures


Similar articles
-
Structure and regulation of the chromatin remodeller ISWI.Nature. 2016 Dec 15;540(7633):466-469. doi: 10.1038/nature20590. Epub 2016 Dec 5. Nature. 2016. PMID: 27919072
-
No need for a power stroke in ISWI-mediated nucleosome sliding.EMBO Rep. 2013 Dec;14(12):1092-7. doi: 10.1038/embor.2013.160. Epub 2013 Oct 11. EMBO Rep. 2013. PMID: 24113208 Free PMC article.
-
Histone H4 tail mediates allosteric regulation of nucleosome remodelling by linker DNA.Nature. 2014 Aug 14;512(7513):213-7. doi: 10.1038/nature13380. Epub 2014 Jun 29. Nature. 2014. PMID: 25043036 Free PMC article.
-
ISWI chromatin remodeling: one primary actor or a coordinated effort?Curr Opin Struct Biol. 2014 Feb;24:150-5. doi: 10.1016/j.sbi.2014.01.010. Epub 2014 Feb 19. Curr Opin Struct Biol. 2014. PMID: 24561830 Free PMC article. Review.
-
Regulation of ISWI chromatin remodelling activity.Chromosoma. 2014 Mar;123(1-2):91-102. doi: 10.1007/s00412-013-0447-4. Epub 2014 Jan 12. Chromosoma. 2014. PMID: 24414837 Review.
Cited by
-
The ACF chromatin-remodeling complex is essential for Polycomb repression.Elife. 2022 Mar 8;11:e77595. doi: 10.7554/eLife.77595. Elife. 2022. PMID: 35257662 Free PMC article.
-
Nucleosome sliding mechanisms: new twists in a looped history.Nat Struct Mol Biol. 2013 Sep;20(9):1026-32. doi: 10.1038/nsmb.2648. Nat Struct Mol Biol. 2013. PMID: 24008565 Review.
-
Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression.Nat Immunol. 2017 May;18(5):499-508. doi: 10.1038/ni.3712. Epub 2017 Mar 20. Nat Immunol. 2017. PMID: 28319097
-
Structural basis of human CHD1 nucleosome recruitment and pausing.Mol Cell. 2025 May 15;85(10):1938-1951.e6. doi: 10.1016/j.molcel.2025.04.020. Epub 2025 May 6. Mol Cell. 2025. PMID: 40334658
-
A nucleotide-driven switch regulates flanking DNA length sensing by a dimeric chromatin remodeler.Mol Cell. 2015 Mar 5;57(5):850-859. doi: 10.1016/j.molcel.2015.01.008. Epub 2015 Feb 12. Mol Cell. 2015. PMID: 25684208 Free PMC article.
References
-
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annual review of biochemistry. 2009;78:273–304. - PubMed
-
- Corona DF, Tamkun JW. Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochim Biophys Acta. 2004;1677:113–119. - PubMed
-
- Zofall M, Persinger J, Kassabov SR, Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol. 2006;13:339–346. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

