Interactions between the neuromodulatory systems and the amygdala: exploratory survey using the Allen Mouse Brain Atlas
- PMID: 23143393
- PMCID: PMC3825589
- DOI: 10.1007/s00429-012-0473-7
Interactions between the neuromodulatory systems and the amygdala: exploratory survey using the Allen Mouse Brain Atlas
Abstract
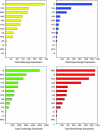
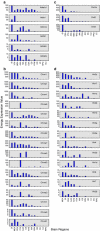
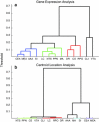
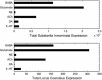
Neuromodulatory systems originate in nuclei localized in the subcortical region of the brain and control fundamental behaviors by interacting with many areas of the central nervous system. An exploratory survey of the cholinergic, dopaminergic, noradrenergic, and serotonergic receptor expression energy in the amygdala, and in the neuromodulatory areas themselves was undertaken using the Allen Mouse Brain Atlas. The amygdala was chosen because of its importance in cognitive behavior and its bidirectional interaction with the neuromodulatory systems. The gene expression data of 38 neuromodulatory receptor subtypes were examined across 13 brain regions. The substantia innominata of the basal forebrain and regions of the amygdala had the highest amount of receptor expression energy for all four neuromodulatory systems examined. The ventral tegmental area also displayed high receptor expression of all four neuromodulators. In contrast, the locus coeruleus displayed low receptor expression energy overall. In general, cholinergic receptor expression was an order of magnitude greater than other neuromodulatory receptors. Since the nuclei of these neuromodulatory systems are thought to be the source of specific neurotransmitters, the projections from these nuclei to target regions may be inferred by receptor expression energy. The comprehensive analysis revealed many connectivity relations and receptor localization that had not been previously reported. The methodology presented here may be applied to other neural systems with similar characteristics, and to other animal models as these brain atlases become available.
Figures




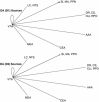


References
-
- Barnes N, Sharp T. A review of central 5-HT receptors and their function. Neuropharmacology. 1999;38(8):1083–1152. - PubMed
-
- Batagelj V, Mrvar A. Pajek-program for large network analysis. Connections. 1998;21(2):47–57.
-
- Baxter MG, Chiba AA. Cognitive functions of the basal forebrain. Curr Opin Neurobiol. 1999;9(2):178–183. - PubMed
-
- Berridge CW, Waterhouse BD. The locus coeruleus-noradrenergic system: modulation of behavioral state and state-dependent cognitive processes. Brain Res Rev. 2003;42(1):33–84. - PubMed
-
- Bhatia SC, Saha S, Manchanda SK, Nayar U. Role of midbrain ventro-lateral tegmental area (VTA) adrenergic mechanisms in facilitation of hypothalamically-induced predatory attack behaviour. Indian J Exp Biol. 1997;35(4):332–337. - PubMed

