Diverse types of genetic variation converge on functional gene networks involved in schizophrenia
- PMID: 23143521
- PMCID: PMC3689007
- DOI: 10.1038/nn.3261
Diverse types of genetic variation converge on functional gene networks involved in schizophrenia
Abstract
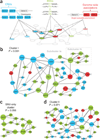
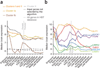
Despite the successful identification of several relevant genomic loci, the underlying molecular mechanisms of schizophrenia remain largely unclear. We developed a computational approach (NETBAG+) that allows an integrated analysis of diverse disease-related genetic data using a unified statistical framework. The application of this approach to schizophrenia-associated genetic variations, obtained using unbiased whole-genome methods, allowed us to identify several cohesive gene networks related to axon guidance, neuronal cell mobility, synaptic function and chromosomal remodeling. The genes forming the networks are highly expressed in the brain, with higher brain expression during prenatal development. The identified networks are functionally related to genes previously implicated in schizophrenia, autism and intellectual disability. A comparative analysis of copy number variants associated with autism and schizophrenia suggests that although the molecular networks implicated in these distinct disorders may be related, the mutations associated with each disease are likely to lead, at least on average, to different functional consequences.
Figures

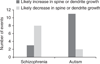

References
-
- O’Donovan MC, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat. Genet. 2008;40:1053–1055. - PubMed
-
- Yue WH, et al. Genome-wide association study identifies a susceptibility locus for schizophrenia in Han Chinese at 11p11.2. Nat. Genet. 2011;43:1228–1231. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

