An extremely halophilic proteobacterium combines a highly acidic proteome with a low cytoplasmic potassium content
- PMID: 23144460
- PMCID: PMC3537055
- DOI: 10.1074/jbc.M112.420505
An extremely halophilic proteobacterium combines a highly acidic proteome with a low cytoplasmic potassium content
Abstract
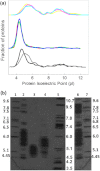
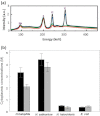
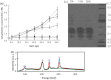
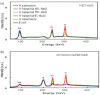
Halophilic archaea accumulate molar concentrations of KCl in their cytoplasm as an osmoprotectant and have evolved highly acidic proteomes that function only at high salinity. We examined osmoprotection in the photosynthetic Proteobacteria Halorhodospira halophila and Halorhodospira halochloris. Genome sequencing and isoelectric focusing gel electrophoresis showed that the proteome of H. halophila is acidic. In line with this finding, H. halophila accumulated molar concentrations of KCl when grown in high salt medium as detected by x-ray microanalysis and plasma emission spectrometry. This result extends the taxonomic range of organisms using KCl as a main osmoprotectant to the Proteobacteria. The closely related organism H. halochloris does not exhibit an acidic proteome, matching its inability to accumulate K(+). This observation indicates recent evolutionary changes in the osmoprotection strategy of these organisms. Upon growth of H. halophila in low salt medium, its cytoplasmic K(+) content matches that of Escherichia coli, revealing an acidic proteome that can function in the absence of high cytoplasmic salt concentrations. These findings necessitate a reassessment of two central aspects of theories for understanding extreme halophiles. First, we conclude that proteome acidity is not driven by stabilizing interactions between K(+) ions and acidic side chains but by the need for maintaining sufficient solvation and hydration of the protein surface at high salinity through strongly hydrated carboxylates. Second, we propose that obligate protein halophilicity is a non-adaptive property resulting from genetic drift in which constructive neutral evolution progressively incorporates weakly stabilizing K(+)-binding sites on an increasingly acidic protein surface.
Figures

Similar articles
-
A potassium chloride to glycine betaine osmoprotectant switch in the extreme halophile Halorhodospira halophila.Sci Rep. 2020 Feb 25;10(1):3383. doi: 10.1038/s41598-020-59231-9. Sci Rep. 2020. PMID: 32098991 Free PMC article.
-
Life at high salt concentrations, intracellular KCl concentrations, and acidic proteomes.Front Microbiol. 2013 Nov 5;4:315. doi: 10.3389/fmicb.2013.00315. Front Microbiol. 2013. PMID: 24204364 Free PMC article. Review.
-
Microbial life at high salt concentrations: phylogenetic and metabolic diversity.Saline Syst. 2008 Apr 15;4:2. doi: 10.1186/1746-1448-4-2. Saline Syst. 2008. PMID: 18412960 Free PMC article.
-
Proteins maintain hydration at high [KCl] concentration regardless of content in acidic amino acids.Biophys J. 2021 Jul 6;120(13):2746-2762. doi: 10.1016/j.bpj.2021.05.015. Epub 2021 Jun 2. Biophys J. 2021. PMID: 34087206 Free PMC article.
-
Halophiles and their enzymes: negativity put to good use.Curr Opin Microbiol. 2015 Jun;25:120-6. doi: 10.1016/j.mib.2015.05.009. Epub 2015 Jun 9. Curr Opin Microbiol. 2015. PMID: 26066288 Free PMC article. Review.
Cited by
-
Methanogenesis and Salt Tolerance Genes of a Novel Halophilic Methanosarcinaceae Metagenome-Assembled Genome from a Former Solar Saltern.Genes (Basel). 2021 Oct 13;12(10):1609. doi: 10.3390/genes12101609. Genes (Basel). 2021. PMID: 34681003 Free PMC article.
-
Salt resistance genes revealed by functional metagenomics from brines and moderate-salinity rhizosphere within a hypersaline environment.Front Microbiol. 2015 Oct 13;6:1121. doi: 10.3389/fmicb.2015.01121. eCollection 2015. Front Microbiol. 2015. PMID: 26528268 Free PMC article.
-
Hans Georg Trüper (1936-2016) and His Contributions to Halophile Research.Life (Basel). 2016 May 12;6(2):19. doi: 10.3390/life6020019. Life (Basel). 2016. PMID: 27187481 Free PMC article.
-
New Halonotius Species Provide Genomics-Based Insights Into Cobalamin Synthesis in Haloarchaea.Front Microbiol. 2019 Aug 27;10:1928. doi: 10.3389/fmicb.2019.01928. eCollection 2019. Front Microbiol. 2019. PMID: 31507553 Free PMC article.
-
Pyrophosphate hydrolysis in the extremely halophilic archaeon Haloarcula japonica is catalyzed by a single enzyme with a broad ionic strength range.Extremophiles. 2017 May;21(3):471-477. doi: 10.1007/s00792-017-0917-3. Epub 2017 Feb 17. Extremophiles. 2017. PMID: 28213825
References
-
- Jackson R. B., Carpenter S. R., Dahm C. N., McKnight D. M., Naiman R. J., Postel S. L., Running S. W. (2001) Water in a changing world. Ecol. Appl. 11, 1027–1045
-
- McGenity T. J., Gemmell R. T., Grant W. D., Stan-Lotter H. (2000) Origins of halophilic microorganisms in ancient salt deposits. Environ. Microbiol. 2, 243–250 - PubMed
-
- Raymond J. C., Sistrom W. R. (1969) Ectothiorhodospira halophila–a new species of genus Ectothiorhodospira. Arch. Mikrobiol. 69, 121–126 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

