Mapping the phosphoproteome of influenza A and B viruses by mass spectrometry
- PMID: 23144613
- PMCID: PMC3493474
- DOI: 10.1371/journal.ppat.1002993
Mapping the phosphoproteome of influenza A and B viruses by mass spectrometry
Abstract
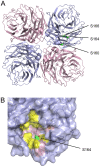
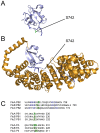
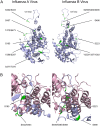
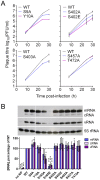
Protein phosphorylation is a common post-translational modification in eukaryotic cells and has a wide range of functional effects. Here, we used mass spectrometry to search for phosphorylated residues in all the proteins of influenza A and B viruses--to the best of our knowledge, the first time such a comprehensive approach has been applied to a virus. We identified 36 novel phosphorylation sites, as well as confirming 3 previously-identified sites. N-terminal processing and ubiquitination of viral proteins was also detected. Phosphorylation was detected in the polymerase proteins (PB2, PB1 and PA), glycoproteins (HA and NA), nucleoprotein (NP), matrix protein (M1), ion channel (M2), non-structural protein (NS1) and nuclear export protein (NEP). Many of the phosphorylation sites detected were conserved between influenza virus genera, indicating the fundamental importance of phosphorylation for all influenza viruses. Their structural context indicates roles for phosphorylation in regulating viral entry and exit (HA and NA); nuclear localisation (PB2, M1, NP, NS1 and, through NP and NEP, of the viral RNA genome); and protein multimerisation (NS1 dimers, M2 tetramers and NP oligomers). Using reverse genetics we show that for NP of influenza A viruses phosphorylation sites in the N-terminal NLS are important for viral growth, whereas mutating sites in the C-terminus has little or no effect. Mutating phosphorylation sites in the oligomerisation domains of NP inhibits viral growth and in some cases transcription and replication of the viral RNA genome. However, constitutive phosphorylation of these sites is not optimal. Taken together, the conservation, structural context and functional significance of phosphorylation sites implies a key role for phosphorylation in influenza biology. By identifying phosphorylation sites throughout the proteomes of influenza A and B viruses we provide a framework for further study of phosphorylation events in the viral life cycle and suggest a range of potential antiviral targets.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Phosphorylation of S-S-S Motif in Nuclear Export Protein (NEP) Plays a Critical Role in Viral Ribonucleoprotein (vRNP) Nuclear Export of Influenza A and B Viruses.Adv Sci (Weinh). 2025 Jan;12(2):e2309477. doi: 10.1002/advs.202309477. Epub 2024 Nov 22. Adv Sci (Weinh). 2025. PMID: 39575547 Free PMC article.
-
The N terminus of the influenza B virus nucleoprotein is essential for virus viability, nuclear localization, and optimal transcription and replication of the viral genome.J Virol. 2014 Nov;88(21):12326-38. doi: 10.1128/JVI.01542-14. Epub 2014 Aug 13. J Virol. 2014. PMID: 25122787 Free PMC article.
-
Phosphoproteome Analysis of Cells Infected with Adapted and Nonadapted Influenza A Virus Reveals Novel Pro- and Antiviral Signaling Networks.J Virol. 2019 Jun 14;93(13):e00528-19. doi: 10.1128/JVI.00528-19. Print 2019 Jul 1. J Virol. 2019. PMID: 30996098 Free PMC article.
-
[Influenza virus genome structure and encoded proteins].Nihon Rinsho. 1997 Oct;55(10):2542-6. Nihon Rinsho. 1997. PMID: 9360369 Review. Japanese.
-
Palmitoylation of influenza virus proteins.Biochem Soc Trans. 2013 Feb 1;41(1):50-5. doi: 10.1042/BST20120210. Biochem Soc Trans. 2013. PMID: 23356257 Review.
Cited by
-
Dissecting the mechanism of signaling-triggered nuclear export of newly synthesized influenza virus ribonucleoprotein complexes.Proc Natl Acad Sci U S A. 2020 Jul 14;117(28):16557-16566. doi: 10.1073/pnas.2002828117. Epub 2020 Jun 29. Proc Natl Acad Sci U S A. 2020. PMID: 32601201 Free PMC article.
-
Cellular Protein Phosphatase 2A Regulates Cell Survival Mechanisms in Influenza A Virus Infection.Int J Mol Sci. 2021 Oct 16;22(20):11164. doi: 10.3390/ijms222011164. Int J Mol Sci. 2021. PMID: 34681823 Free PMC article.
-
Influenza Virus Infections and Cellular Kinases.Viruses. 2019 Feb 20;11(2):171. doi: 10.3390/v11020171. Viruses. 2019. PMID: 30791550 Free PMC article. Review.
-
Experimental Approaches to Identify Host Factors Important for Influenza Virus.Cold Spring Harb Perspect Med. 2020 Dec 1;10(12):a038521. doi: 10.1101/cshperspect.a038521. Cold Spring Harb Perspect Med. 2020. PMID: 31871241 Free PMC article. Review.
-
Functional Constraint Profiling of a Viral Protein Reveals Discordance of Evolutionary Conservation and Functionality.PLoS Genet. 2015 Jul 1;11(7):e1005310. doi: 10.1371/journal.pgen.1005310. eCollection 2015 Jul. PLoS Genet. 2015. PMID: 26132554 Free PMC article.
References
-
- Lin YP, Gregory V, Bennett M, Hay A (2004) Recent changes among human influenza viruses. Virus Res 103: 47–52. - PubMed
-
- Buonagurio DA, Nakada S, Fitch WM, Palese P (1986) Epidemiology of influenza C virus in man: multiple evolutionary lineages and low rate of change. Virology 153: 12–21. - PubMed
-
- Cohen P (2000) The regulation of protein function by multisite phosphorylation–a 25 year update. Trends Biochem Sci 25: 596–601. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

