Microhomology directs diverse DNA break repair pathways and chromosomal translocations
- PMID: 23144625
- PMCID: PMC3493447
- DOI: 10.1371/journal.pgen.1003026
Microhomology directs diverse DNA break repair pathways and chromosomal translocations
Abstract
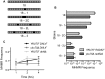
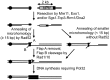
Chromosomal structural change triggers carcinogenesis and the formation of other genetic diseases. The breakpoint junctions of these rearrangements often contain small overlapping sequences called "microhomology," yet the genetic pathway(s) responsible have yet to be defined. We report a simple genetic system to detect microhomology-mediated repair (MHMR) events after a DNA double-strand break (DSB) in budding yeast cells. MHMR using >15 bp operates as a single-strand annealing variant, requiring the non-essential DNA polymerase subunit Pol32. MHMR is inhibited by sequence mismatches, but independent of extensive DNA synthesis like break-induced replication. However, MHMR using less than 14 bp is genetically distinct from that using longer microhomology and far less efficient for the repair of distant DSBs. MHMR catalyzes chromosomal translocation almost as efficiently as intra-chromosomal repair. The results suggest that the intrinsic annealing propensity between microhomology sequences efficiently leads to chromosomal rearrangements.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

