Mechanisms employed by Escherichia coli to prevent ribonucleotide incorporation into genomic DNA by Pol V
- PMID: 23144626
- PMCID: PMC3493448
- DOI: 10.1371/journal.pgen.1003030
Mechanisms employed by Escherichia coli to prevent ribonucleotide incorporation into genomic DNA by Pol V
Abstract
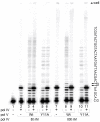
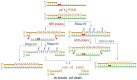
Escherichia coli pol V (UmuD'(2)C), the main translesion DNA polymerase, ensures continued nascent strand extension when the cellular replicase is blocked by unrepaired DNA lesions. Pol V is characterized by low sugar selectivity, which can be further reduced by a Y11A "steric-gate" substitution in UmuC that enables pol V to preferentially incorporate rNTPs over dNTPs in vitro. Despite efficient error-prone translesion synthesis catalyzed by UmuC_Y11A in vitro, strains expressing umuC_Y11A exhibit low UV mutability and UV resistance. Here, we show that these phenotypes result from the concomitant dual actions of Ribonuclease HII (RNase HII) initiating removal of rNMPs from the nascent DNA strand and nucleotide excision repair (NER) removing UV lesions from the parental strand. In the absence of either repair pathway, UV resistance and mutagenesis conferred by umuC_Y11A is significantly enhanced, suggesting that the combined actions of RNase HII and NER lead to double-strand breaks that result in reduced cell viability. We present evidence that the Y11A-specific UV phenotype is tempered by pol IV in vivo. At physiological ratios of the two polymerases, pol IV inhibits pol V-catalyzed translesion synthesis (TLS) past UV lesions and significantly reduces the number of Y11A-incorporated rNTPs by limiting the length of the pol V-dependent TLS tract generated during lesion bypass in vitro. In a recA730 lexA(Def) ΔumuDC ΔdinB strain, plasmid-encoded wild-type pol V promotes high levels of spontaneous mutagenesis. However, umuC_Y11A-dependent spontaneous mutagenesis is only ~7% of that observed with wild-type pol V, but increases to ~39% of wild-type levels in an isogenic ΔrnhB strain and ~72% of wild-type levels in a ΔrnhA ΔrnhB double mutant. Our observations suggest that errant ribonucleotides incorporated by pol V can be tolerated in the E. coli genome, but at the cost of higher levels of cellular mutagenesis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Removal of misincorporated ribonucleotides from prokaryotic genomes: an unexpected role for nucleotide excision repair.PLoS Genet. 2013 Nov;9(11):e1003878. doi: 10.1371/journal.pgen.1003878. Epub 2013 Nov 7. PLoS Genet. 2013. PMID: 24244177 Free PMC article.
-
Investigating the mechanisms of ribonucleotide excision repair in Escherichia coli.Mutat Res. 2014 Mar;761:21-33. doi: 10.1016/j.mrfmmm.2014.01.005. Epub 2014 Feb 1. Mutat Res. 2014. PMID: 24495324 Free PMC article.
-
Escherichia coli UmuC active site mutants: effects on translesion DNA synthesis, mutagenesis and cell survival.DNA Repair (Amst). 2012 Sep 1;11(9):726-32. doi: 10.1016/j.dnarep.2012.06.005. Epub 2012 Jul 10. DNA Repair (Amst). 2012. PMID: 22784977 Free PMC article.
-
A Comprehensive View of Translesion Synthesis in Escherichia coli.Microbiol Mol Biol Rev. 2020 Jun 17;84(3):e00002-20. doi: 10.1128/MMBR.00002-20. Print 2020 Aug 19. Microbiol Mol Biol Rev. 2020. PMID: 32554755 Free PMC article. Review.
-
Mutations for Worse or Better: Low-Fidelity DNA Synthesis by SOS DNA Polymerase V Is a Tightly Regulated Double-Edged Sword.Biochemistry. 2016 Apr 26;55(16):2309-18. doi: 10.1021/acs.biochem.6b00117. Epub 2016 Apr 12. Biochemistry. 2016. PMID: 27043933 Free PMC article. Review.
Cited by
-
Ribonucleotides in bacterial DNA.Crit Rev Biochem Mol Biol. 2015;50(3):181-93. doi: 10.3109/10409238.2014.981647. Epub 2014 Nov 12. Crit Rev Biochem Mol Biol. 2015. PMID: 25387798 Free PMC article. Review.
-
Substrate Specificity for Bacterial RNases HII and HIII Is Influenced by Metal Availability.J Bacteriol. 2018 Jan 24;200(4):e00401-17. doi: 10.1128/JB.00401-17. Print 2018 Feb 15. J Bacteriol. 2018. PMID: 29084857 Free PMC article.
-
"Life is short, and art is long": RNA degradation in cyanobacteria and model bacteria.mLife. 2022 Mar 24;1(1):21-39. doi: 10.1002/mlf2.12015. eCollection 2022 Mar. mLife. 2022. PMID: 38818322 Free PMC article. Review.
-
CroSR391 , an ortholog of the λ Cro repressor, plays a major role in suppressing polVR391 -dependent mutagenesis.Mol Microbiol. 2021 Sep;116(3):877-889. doi: 10.1111/mmi.14777. Epub 2021 Jul 12. Mol Microbiol. 2021. PMID: 34184328 Free PMC article.
-
Removal of misincorporated ribonucleotides from prokaryotic genomes: an unexpected role for nucleotide excision repair.PLoS Genet. 2013 Nov;9(11):e1003878. doi: 10.1371/journal.pgen.1003878. Epub 2013 Nov 7. PLoS Genet. 2013. PMID: 24244177 Free PMC article.
References
-
- Vaisman A, McDonald JP, Woodgate R (2012) Translesion DNA Synthesis. In: Böck A, Curtiss R, Kaper JB, Karp PD, Neidhardt FC et al.., editors. EcoSal: Escherichia coli and Salmonella: cellular and molecular miology. Washington, DC: American Society for Biology.
-
- Cai H, Yu H, McEntee K, Kunkel TA, Goodman MF (1995) Purification and properties of wild-type and exonuclease-deficient DNA polymerase II from Escherichia coli . J Biol Chem 270: 15327–15335. - PubMed
-
- Ohmori H, Hatada E, Qiao Y, Tsuji M, Fukuda R (1995) dinP, a new gene in Escherichia coli, whose product shows similarities to UmuC and its homologues. Mut Res 347: 1–7. - PubMed
-
- Wagner J, Gruz P, Kim SR, Yamada M, Matsui K, et al. (1999) The dinB gene encodes an novel Escherichia coli DNA polymerase (DNA pol IV) involved in mutagenesis. Mol Cell 4: 281–286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

