Informed conditioning on clinical covariates increases power in case-control association studies
- PMID: 23144628
- PMCID: PMC3493452
- DOI: 10.1371/journal.pgen.1003032
Informed conditioning on clinical covariates increases power in case-control association studies
Abstract
Genetic case-control association studies often include data on clinical covariates, such as body mass index (BMI), smoking status, or age, that may modify the underlying genetic risk of case or control samples. For example, in type 2 diabetes, odds ratios for established variants estimated from low-BMI cases are larger than those estimated from high-BMI cases. An unanswered question is how to use this information to maximize statistical power in case-control studies that ascertain individuals on the basis of phenotype (case-control ascertainment) or phenotype and clinical covariates (case-control-covariate ascertainment). While current approaches improve power in studies with random ascertainment, they often lose power under case-control ascertainment and fail to capture available power increases under case-control-covariate ascertainment. We show that an informed conditioning approach, based on the liability threshold model with parameters informed by external epidemiological information, fully accounts for disease prevalence and non-random ascertainment of phenotype as well as covariates and provides a substantial increase in power while maintaining a properly controlled false-positive rate. Our method outperforms standard case-control association tests with or without covariates, tests of gene x covariate interaction, and previously proposed tests for dealing with covariates in ascertained data, with especially large improvements in the case of case-control-covariate ascertainment. We investigate empirical case-control studies of type 2 diabetes, prostate cancer, lung cancer, breast cancer, rheumatoid arthritis, age-related macular degeneration, and end-stage kidney disease over a total of 89,726 samples. In these datasets, informed conditioning outperforms logistic regression for 115 of the 157 known associated variants investigated (P-value = 1 × 10(-9)). The improvement varied across diseases with a 16% median increase in χ(2) test statistics and a commensurate increase in power. This suggests that applying our method to existing and future association studies of these diseases may identify novel disease loci.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

 )+m. High-BMI cases have a lower posterior mean relative to low-BMI cases since they require a smaller contribution from genetics to exceed the threshold in the liability threshold model.
)+m. High-BMI cases have a lower posterior mean relative to low-BMI cases since they require a smaller contribution from genetics to exceed the threshold in the liability threshold model.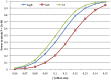
References
-
- Ellis KL, Pilbrow AP, Frampton CM, Doughty RN, Whalley GA, et al. (2010) A common variant at chromosome 9P21.3 is associated with age of onset of coronary disease but not subsequent mortality. Circ Cardiovasc Genet 3: 286–293. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 CA092824/CA/NCI NIH HHS/United States
- HL043851/HL/NHLBI NIH HHS/United States
- U01-CA98233-07/CA/NCI NIH HHS/United States
- R01 HG006399/HG/NHGRI NIH HHS/United States
- U01-CA98216-06/CA/NCI NIH HHS/United States
- R01 CA047988/CA/NCI NIH HHS/United States
- U19 HL069757/HL/NHLBI NIH HHS/United States
- U01 CA098758/CA/NCI NIH HHS/United States
- U01-CA98758-07/CA/NCI NIH HHS/United States
- R21 ES020754/ES/NIEHS NIH HHS/United States
- U01 HL069757/HL/NHLBI NIH HHS/United States
- U01 CA098710/CA/NCI NIH HHS/United States
- U19 CA148127/CA/NCI NIH HHS/United States
- R01 ES020337/ES/NIEHS NIH HHS/United States
- T32 ES007142/ES/NIEHS NIH HHS/United States
- HL69757/HL/NHLBI NIH HHS/United States
- R01 HL043851/HL/NHLBI NIH HHS/United States
- 5T32ES007142-27/ES/NIEHS NIH HHS/United States
- 17552/ARC_/Arthritis Research UK/United Kingdom
- U01-CA98710-06/CA/NCI NIH HHS/United States
- U01 CA098216/CA/NCI NIH HHS/United States
- CA 047988/CA/NCI NIH HHS/United States
- U01 CA098233/CA/NCI NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

