The success of acinetobacter species; genetic, metabolic and virulence attributes
- PMID: 23144699
- PMCID: PMC3483291
- DOI: 10.1371/journal.pone.0046984
The success of acinetobacter species; genetic, metabolic and virulence attributes
Abstract
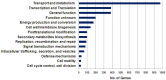
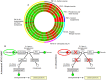
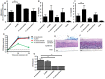
An understanding of why certain Acinetobacter species are more successful in causing nosocomial infections, transmission and epidemic spread in healthcare institutions compared with other species is lacking. We used genomic, phenotypic and virulence studies to identify differences between Acinetobacter species. Fourteen strains representing nine species were examined. Genomic analysis of six strains showed that the A. baumannii core genome contains many genes important for diverse metabolism and survival in the host. Most of the A. baumannii core genes were also present in one or more of the less clinically successful species. In contrast, when the accessory genome of an individual A. baumannii strain was compared to a strain of a less successful species (A. calcoaceticus RUH2202), many operons with putative virulence function were found to be present only in the A. baumannii strain, including the csu operon, the acinetobactin chromosomal cluster, and bacterial defence mechanisms. Phenotype microarray analysis showed that compared to A. calcoaceticus (RUH2202), A. baumannii ATCC 19606(T) was able to utilise nitrogen sources more effectively and was more tolerant to pH, osmotic and antimicrobial stress. Virulence differences were also observed, with A. baumannii ATCC 19606(T), A. pittii SH024, and A. nosocomialis RUH2624 persisting and forming larger biofilms on human skin than A. calcoaceticus. A. baumannii ATCC 19606(T) and A. pittii SH024 were also able to survive in a murine thigh infection model, whereas the other two species were eradicated. The current study provides important insights into the elucidation of differences in clinical relevance among Acinetobacter species.
Conflict of interest statement
Figures


References
-
- Dijkshoorn L, Nemec A, Seifert H (2007) An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol 5: 939–951. - PubMed
-
- Nemec A, Krizova L, Maixnerova M, van der Reijden TJ, Deschaght P, et al. (2011) Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU). Res Microbiol 162: 393–404. - PubMed
-
- Chuang YC, Sheng WH, Li SY, Lin YC, Wang JT, et al. (2011) Influence of genospecies of Acinetobacter baumannii complex on clinical outcomes of patients with acinetobacter bacteremia. Clin Infect Dis 52: 352–360. - PubMed
-
- Koh TH, Tan TT, Khoo CT, Ng SY, Tan TY, et al. (2012) Acinetobacter calcoaceticus- Acinetobacter baumannii complex species in clinical specimens in Singapore. Epidemiol Infect 1–4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

