In vivo capsular switch in Streptococcus pneumoniae--analysis by whole genome sequencing
- PMID: 23144841
- PMCID: PMC3493582
- DOI: 10.1371/journal.pone.0047983
In vivo capsular switch in Streptococcus pneumoniae--analysis by whole genome sequencing
Abstract
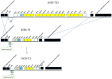
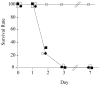
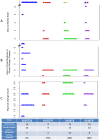
Two multidrug resistant strains of Streptococcus pneumoniae - SV35-T23 (capsular type 23F) and SV36-T3 (capsular type 3) were recovered from the nasopharynx of two adult patients during an outbreak of pneumococcal disease in a New York hospital in 1996. Both strains belonged to the pandemic lineage PMEN1 but they differed strikingly in virulence when tested in the mouse model of IP infection: as few as 1000 CFU of SV36 killed all mice within 24 hours after inoculation while SV35-T23 was avirulent.Whole genome sequencing (WGS) of the two isolates was performed (i) to test if these two isolates belonging to the same clonal type and recovered from an identical epidemiological scenario only differed in their capsular genes? and (ii) to test if the vast difference in virulence between the strains was mostly - or exclusively - due to the type III capsule. WGS demonstrated extensive differences between the two isolates including over 2500 single nucleotide polymorphisms in core genes and also differences in 36 genetic determinants: 25 of which were unique to SV35-T23 and 11 unique to strain SV36-T3. Nineteen of these differences were capsular genes and 9 bacteriocin genes.Using genetic transformation in the laboratory, the capsular region of SV35-T23 was replaced by the type 3 capsular genes from SV36-T3 to generate the recombinant SV35-T3* which was as virulent as the parental strain SV36-T3* in the murine model and the type 3 capsule was the major virulence factor in the chinchilla model as well. On the other hand, a careful comparison of strains SV36-T3 and the laboratory constructed SV35-T3* in the chinchilla model suggested that some additional determinants present in SV36 but not in the laboratory recombinant may also contribute to the progression of middle ear disease. The nature of this determinants remains to be identified.
Conflict of interest statement
Figures


References
-
- Aniansson G, Alm B, Andersson B, Larsson P, Nylén O, et al. (1992) Nasopharyngeal colonization during the first year of life. J Infect Dis 165 Suppl 1: S38–42. - PubMed
-
- Austrian R (1986) Some aspects of the pneumococcal carrier state. J Antimicrob Chemother 18 Suppl A: 35–45. - PubMed
-
- Gray BM, Converse GM, Dillon HC (1980) Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage, and infection during the first 24 months of life. J Infect Dis 142: 923–933. - PubMed
-
- Bogaert D, De Groot R, Hermans PW (2004) Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis 4: 144–154. - PubMed
-
- World Health Organizaiton website. Streptococcus pneumoniae 2008; Available: http://www.who.int/vaccine_research/diseases/ari/en/index5.html. Accessed 2012 September 5.
Publication types
MeSH terms
Grants and funding
- C76HF03009/PHS HHS/United States
- R01 DC002148/DC/NIDCD NIH HHS/United States
- DC02148 S1/DC/NIDCD NIH HHS/United States
- C76HF09819/PHS HHS/United States
- DC05659/DC/NIDCD NIH HHS/United States
- R01 AI080935/AI/NIAID NIH HHS/United States
- 1R01AI080935-01A1/AI/NIAID NIH HHS/United States
- C76HF00659/PHS HHS/United States
- R01 DC004173/DC/NIDCD NIH HHS/United States
- AI080935/AI/NIAID NIH HHS/United States
- SFRH/BD/30103/2006/BD/FDA HHS/United States
- R01 DC005659/DC/NIDCD NIH HHS/United States
- DC04173/DC/NIDCD NIH HHS/United States
- DC02148/DC/NIDCD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

