Integrative analysis reveals relationships of genetic and epigenetic alterations in osteosarcoma
- PMID: 23144859
- PMCID: PMC3492335
- DOI: 10.1371/journal.pone.0048262
Integrative analysis reveals relationships of genetic and epigenetic alterations in osteosarcoma
Abstract
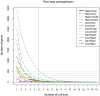
Background: Osteosarcomas are the most common non-haematological primary malignant tumours of bone, and all conventional osteosarcomas are high-grade tumours showing complex genomic aberrations. We have integrated genome-wide genetic and epigenetic profiles from the EuroBoNeT panel of 19 human osteosarcoma cell lines based on microarray technologies.
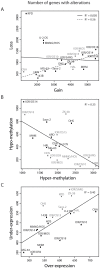
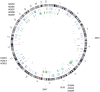
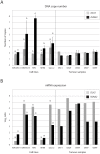
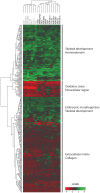
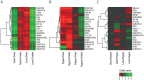
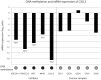
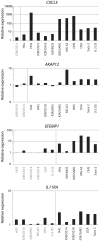
Principal findings: The cell lines showed complex patterns of DNA copy number changes, where genomic copy number gains were significantly associated with gene-rich regions and losses with gene-poor regions. By integrating the datasets, 350 genes were identified as having two types of aberrations (gain/over-expression, hypo-methylation/over-expression, loss/under-expression or hyper-methylation/under-expression) using a recurrence threshold of 6/19 (>30%) cell lines. The genes showed in general alterations in either DNA copy number or DNA methylation, both within individual samples and across the sample panel. These 350 genes are involved in embryonic skeletal system development and morphogenesis, as well as remodelling of extracellular matrix. The aberrations of three selected genes, CXCL5, DLX5 and RUNX2, were validated in five cell lines and five tumour samples using PCR techniques. Several genes were hyper-methylated and under-expressed compared to normal osteoblasts, and expression could be reactivated by demethylation using 5-Aza-2'-deoxycytidine treatment for four genes tested; AKAP12, CXCL5, EFEMP1 and IL11RA. Globally, there was as expected a significant positive association between gain and over-expression, loss and under-expression as well as hyper-methylation and under-expression, but gain was also associated with hyper-methylation and under-expression, suggesting that hyper-methylation may oppose the effects of increased copy number for detrimental genes.
Conclusions: Integrative analysis of genome-wide genetic and epigenetic alterations identified dependencies and relationships between DNA copy number, DNA methylation and mRNA expression in osteosarcomas, contributing to better understanding of osteosarcoma biology.
Conflict of interest statement
Figures

References
-
- Raymond AK, Ayala AG, Knuutila S (2002) Conventional osteosarcoma. In: Fletcher CDM, Unni KK, Mertens F, editors. World Health Organization Classification of Tumours Pathology and Genetics of Tumours of Soft Tissue and Bone. Lyon: IARC Press.
-
- Aksnes LH, Hall KS, Folleraas G, Stenwig AE, Bjerkehagen B, et al. (2006) Management of high-grade bone sarcomas over two decades: the Norwegian Radium Hospital experience. Acta Oncol 45: 38–46. - PubMed
-
- Hattinger CM, Pasello M, Ferrari S, Picci P, Serra M (2010) Emerging drugs for high-grade osteosarcoma. Expert Opin Emerg Drugs 15: 615–634. - PubMed
-
- Lewis IJ, Nooij MA, Whelan J, Sydes MR, Grimer R, et al. (2007) Improvement in histologic response but not survival in osteosarcoma patients treated with intensified chemotherapy: a randomized phase III trial of the European Osteosarcoma Intergroup. J Natl Cancer Inst 99: 112–128. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

