A molecular phylogeny of Hemiptera inferred from mitochondrial genome sequences
- PMID: 23144967
- PMCID: PMC3493603
- DOI: 10.1371/journal.pone.0048778
A molecular phylogeny of Hemiptera inferred from mitochondrial genome sequences
Abstract
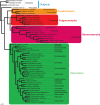
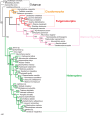
Classically, Hemiptera is comprised of two suborders: Homoptera and Heteroptera. Homoptera includes Cicadomorpha, Fulgoromorpha and Sternorrhyncha. However, according to previous molecular phylogenetic studies based on 18S rDNA, Fulgoromorpha has a closer relationship to Heteroptera than to other hemipterans, leaving Homoptera as paraphyletic. Therefore, the position of Fulgoromorpha is important for studying phylogenetic structure of Hemiptera. We inferred the evolutionary affiliations of twenty-five superfamilies of Hemiptera using mitochondrial protein-coding genes and rRNAs. We sequenced three mitogenomes, from Pyrops candelaria, Lycorma delicatula and Ricania marginalis, representing two additional families in Fulgoromorpha. Pyrops and Lycorma are representatives of an additional major family Fulgoridae in Fulgoromorpha, whereas Ricania is a second representative of the highly derived clade Ricaniidae. The organization and size of these mitogenomes are similar to those of the sequenced fulgoroid species. Our consensus phylogeny of Hemiptera largely supported the relationships (((Fulgoromorpha,Sternorrhyncha),Cicadomorpha),Heteroptera), and thus supported the classic phylogeny of Hemiptera. Selection of optimal evolutionary models (exclusion and inclusion of two rRNA genes or of third codon positions of protein-coding genes) demonstrated that rapidly evolving and saturated sites should be removed from the analyses.
Conflict of interest statement
Figures




Similar articles
-
Characterization, Comparative Analysis and Phylogenetic Implications of Mitogenomes of Fulgoridae (Hemiptera: Fulgoromorpha).Genes (Basel). 2021 Jul 30;12(8):1185. doi: 10.3390/genes12081185. Genes (Basel). 2021. PMID: 34440359 Free PMC article.
-
Characterization of the complete mitochondrial genome of Loxocephala sichuanensis (Hemiptera: Eurybrachidae) with the phylogenetic analyses of Fulgoromorpha.Sci Rep. 2024 Sep 20;14(1):21981. doi: 10.1038/s41598-024-71948-5. Sci Rep. 2024. PMID: 39304689 Free PMC article.
-
Complete mitochondrial genome of the small brown planthopper, Laodelphax striatellus (Delphacidae: Hemiptera), with a novel gene order.Zoolog Sci. 2009 Dec;26(12):851-60. doi: 10.2108/zsj.26.851. Zoolog Sci. 2009. PMID: 19968473
-
Hemipteran mitochondrial genomes: features, structures and implications for phylogeny.Int J Mol Sci. 2015 Jun 1;16(6):12382-404. doi: 10.3390/ijms160612382. Int J Mol Sci. 2015. PMID: 26039239 Free PMC article. Review.
-
True Parthenogenesis and Female-Biased Sex Ratios in Cicadomorpha and Fulgoromorpha (Hemiptera, Auchenorrhyncha).Insects. 2023 Oct 17;14(10):820. doi: 10.3390/insects14100820. Insects. 2023. PMID: 37887832 Free PMC article. Review.
Cited by
-
New Molecular Phylogenetic Evidence Confirms Independent Origin of Coxal Combs in the Families of the 'Cydnoid' Complex (Hemiptera: Heteroptera: Pentatomoidea).Insects. 2024 Oct 11;15(10):792. doi: 10.3390/insects15100792. Insects. 2024. PMID: 39452368 Free PMC article.
-
Chromosomal evolutionary dynamics of four multigene families in Coreidae and Pentatomidae (Heteroptera) true bugs.Mol Genet Genomics. 2016 Oct;291(5):1919-25. doi: 10.1007/s00438-016-1229-5. Epub 2016 Jul 5. Mol Genet Genomics. 2016. PMID: 27380138
-
Rearrangement of mitochondrial tRNA genes in flat bugs (Hemiptera: Aradidae).Sci Rep. 2016 May 16;6:25725. doi: 10.1038/srep25725. Sci Rep. 2016. PMID: 27180804 Free PMC article.
-
Amino acid transporter expansions associated with the evolution of obligate endosymbiosis in sap-feeding insects (Hemiptera: sternorrhyncha).BMC Evol Biol. 2015 Mar 25;15:52. doi: 10.1186/s12862-015-0315-3. BMC Evol Biol. 2015. PMID: 25887093 Free PMC article.
-
Characterization, Comparative Analysis and Phylogenetic Implications of Mitogenomes of Fulgoridae (Hemiptera: Fulgoromorpha).Genes (Basel). 2021 Jul 30;12(8):1185. doi: 10.3390/genes12081185. Genes (Basel). 2021. PMID: 34440359 Free PMC article.
References
-
- Wolstenholme DR (1992) Animal mitochondrial DNA: structure and evolution. Int Rev Cytol 141: 173–216. - PubMed
-
- Inohira K, Hara T, Matsuura ET (1997) Nucleotide sequence divergence in the A+T-rich region of mitochondrial DNA in Drosophila simulans and Drosophila mauritiana . Mol Biol Evol 14: 814–822. - PubMed
-
- Nardi F, Spinsanti G, Boore JL, Carapelli A, Dallai R, et al. (2003) Hexapod origins: monophyletic or paraphyletic? Science 299: 1887–1889. - PubMed
-
- Sheffield NC, Song H, Cameron SL, Whiting MF (2009) Nonstationary Evolution and Compositional Heterogeneity in Beetle Mitochondrial Phylogenomics. Syst Biol 58: 381–394. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

