HERV-E-mediated modulation of PLA2G4A transcription in urothelial carcinoma
- PMID: 23145155
- PMCID: PMC3492278
- DOI: 10.1371/journal.pone.0049341
HERV-E-mediated modulation of PLA2G4A transcription in urothelial carcinoma
Abstract
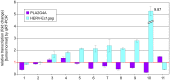
Human endogenous retroviruses (HERV) and related elements account for more than 8% of the human genome and significantly contribute to the human transcriptome by long terminal repeat (LTR) promoter activity. In this context, HERVs are thought to intervene in the expression of adjacent genes by providing regulatory sequences (cis-effect) or via noncoding RNA including natural antisense transcripts. To address the potential impact of HERV activity in urothelial carcinoma, we comparatively analyzed the HERV transcription profiles in paired samples of non-malignant urothelium and urothelial carcinoma derived from 13 patients with bladder cancer by means of a retrovirus-specific microarray (RetroArray). We established a characteristic HERV signature consisting of six ubiquitously active HERV subgroups (E4-1, HERV-Rb, ERV9, HERV-K-T47D, NMWV3, HERV-KC4). The transcription pattern is largely identical in human urothelial carcinoma, non-malignant urothelial tissue, four tumor-derived cell lines and in a non-malignant urothelial cell line (UROtsa). Quantitative reverse transcriptase PCR (qRT-PCR) of HERV-E4-1, HERV-K(HML-6) and HERV-T(S71-TK1) revealed a bias to lower HERV activity in carcinoma samples compared to non-malignant tissue. Determination of active HERV-E4-1 loci by cloning and sequencing revealed six HERV-E4-1 proviral loci that are differentially regulated in urothelial carcinoma cells and normal tissue. Two full-length HERV-E4-1 proviruses, HERV-Ec1 and HERV-Ec6, are located in antisense orientation in introns of the genes PLA2G4A and RNGTT, respectively. PLA2G4A encodes a cytosolic phospholipase A2 (cPLA2) that is dysregulated in many human tumors. PLA2G4A and HERV-Ec1 displayed reciprocal transcript levels in 7 of 11 urothelial carcinoma patients. Moreover, reciprocal shifts were observed after treatment of UROtsa cells with HERV-Ec1 and PLA2G4A-directed siRNAs or 5-aza-2'-deoxycytidine (aza-dC) pointing to an antagonistic regulation of PLA2G4A and HERV-Ec1 transcription in human urothelial cells. We suggest that transcription of HERV-Ec1 contributes to fine tuning of cPLA2 expression, thereby facilitating tumorigenesis.
Conflict of interest statement
Figures





References
-
- Conley AB, Piriyapongsa J, Jordan IK (2008) Retroviral promoters in the human genome. Bioinformatics 24: 1563–1567. - PubMed
-
- Jordan IK, Rogozin IB, Glazko GV, Koonin EV (2003) Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet 19: 68–72. - PubMed
-
- Cohen CJ, Lock WM, Mager DL (2009) Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene 448: 105–114. - PubMed
-
- van de Lagemaat LN, Landry J-R, Mager DL, Medstrand P (2003) Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet 19: 530–536. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

